Abstract
Recent evidences suggested that growth and differentiation of pancreatic cell lineages, including the insulin-producing β-cells, depend on proper tissue-architecture, epithelial remodeling and cell positioning within the branching pancreatic epithelium. We recently found that Rho GTPase and its regulator, Stard13 RhoGAP, coordinate morphogenesis with growth in the developing pancreas. Conditional mutation of Stard13 in the mouse pancreas hampers epithelial remodeling and distal tip domain formation, affecting proliferation and expansion of pancreatic progenitors. These defects eventually result in pancreatic hypoplasia at birth. Stard13 acts by regulating Rho signaling spatially and temporally during pancreas development. In line with this, pharmacological activation or inhibition of Rho mimics or rescues, respectively, the defects observed in Stard13-deficient embryos and pancreatic organ cultures. Furthermore, in the absence of Stard13 uninhibited Rho activity affects the actomyosin contractile network, disrupting its apical distribution and hampering coordinated cell-shape changes. These results unveil therefore the crucial role of actin cytoskeletal dynamics during the onset of pancreatic branching morphogenesis. Finally, our findings define a reciprocal interaction between the actin-MAL/SRF and the MAPK signaling to locally regulate progenitor cell proliferation in the pancreas.
During embryogenesis developing organs acquire their final functional architectures at the same time as their residing cells proliferate and undergo differentiation. This leads to interesting unsolved problems: How is morphogenesis integrated with growth and differentiation events during organ formation? Do they influence each other? For instance, does change in proliferation affect change in shape?
Tissue forms can be the result of spatial differentials in cell proliferation. For example, during epithelial branching morphogenesis and vascular sprouting, cells located at the tips of branching buds generally proliferate more rapidly than neighboring cells located in the ducts.Citation1,Citation2 This so-called “localized cell proliferation” is an important morphogenetic mechanism in kidney development, directing the extension of the epithelial branches.Citation2,Citation3 However, there are examples in which bud outgrowth (e.g., in the lung) has been found to precede localized proliferation, suggesting that alternative mechanisms may be at the origin of shape acquisition.Citation4 Similarly, complete block of proliferation in pancreas organ cultures does not hamper the initiation of primary branches, suggesting that “localized proliferation” is not the only mechanism underlying bud outgrowth and branch formation in the developing pancreas as well.Citation5 Furthermore, these observations underscore another important facet of the problem, which is the control of the relative spatial position of cells within a developing tissue for being correctly formed and functional. It is likely that initial change in shape allocates a certain cell type in a special position (e.g., progenitor cells at the tip of the branch) where it can acquire its identity and proliferate or differentiate, accordingly.
Here, we focus on recent advances in the understanding of how branching morphogenesis is coupled with growth and, possibly, differentiation in the pancreas, emphasizing the role of the Rho-actin signaling in these processes. The pancreas is a mixed exocrine and endocrine gland responsible for vital functions in our body, including the digestion and glucose metabolism.Citation6,Citation7 During embryonic development, the pancreas originates from distinct embryonic outgrowths of the dorsal and ventral primitive gut endoderm at embryonic day (E) 9.5 in the mouse embryo.Citation7,Citation8 Both outgrowths evaginate into the surrounding mesenchyme as solid epithelial buds, which subsequently undergo proliferation, differentiation and morphogenesis and, eventually, fuse together to form the definitive organ.Citation7,Citation8 Recent evidence suggests that pancreatic organ size is constrained by the initial progenitor population.Citation9,Citation10 When pancreatic fate is specified all Pdx1+ epithelial cells appear to act as progenitor cells, whereas, subsequently, at the initiation of branching [between E12 and E14] progenitor cells become confined to the tip of the branches.Citation7,Citation10,Citation11 Indeed, lineage restriction within the pancreatic epithelium delineates a distal tip and a trunk domain of the branches.Citation10 Tip progenitor cells are capable of generating all pancreatic cell types, undergo limited self-renewal and proliferate much faster than the trunk cells.Citation5,Citation10 After E14.5, the tip undergoes a developmental switch and its fate becomes restricted to the exocrine lineage, while most of the trunk cells appear committed to the endocrine lineage.Citation6,Citation7,Citation10 Thus, the branches represent a perfect example of modular unit—in which the tip might serve as an organizing center for pancreatic organogenesis, fueling the tissue with multipotent progenitor cells that divide rapidly and, possibly, in a directional fashion, leaving behind, in the trunk, their more differentiated progeny (e.g., the endocrine and duct cells). In this scenario, lineage restriction needs to be tightly coupled with branching morphogenesis to ultimately build functional tissue-architecture in the pancreas.
Small Rho-GTPases are well-known regulators of many essential cellular processes, including cytoskeletal dynamics, cell polarity, adhesion and migration, and, thereby, control morphogenesis in a variety of epithelial tissues.Citation12,Citation13 Rho, Cdc42 and Rac1, the most studied members of the Rho GTPase family, have been recently shown to control different aspects of pancreatic morphogenesis, including initiation of branching process, tubulogenesis and islet cells migration, respectively.Citation14-Citation16
The cycling of small GTPases between an active (GTP-bound) and inactive (GDP-bound) state is tightly regulated by three classes of proteins: the guanine-nucleotide exchange factors (GEF), GTPases-activating proteins (GAP) and GDP-dissociation inhibitors (GDI).Citation17 Specifically, the GAPs inactivate small GTPases by accelerating their intrinsic GTPase activity and converting them into inactive GDP-bound form.Citation17,Citation18 Because of the numerous roles and ubiquitous nature of small GTPases, preferential tissue expression of these regulatory proteins may be critical for precisely timed and localized activity of Rho family members. In line with this, we recently identified the RhoGAP protein Stard13 as an essential mechanism to spatio-temporally restrict Rho signaling in the embryonic pancreas.Citation16 The loss of this type of control in the Stard13 conditional knockout mouse results into constitutive Rho activation and severe perturbation of pancreas organogenesis ().Citation16
Figure 1. Schematic diagram representing pancreatic branching morphogenesis and lineage restriction in wild-type (upper panel) and Stard13-deficient (bottom panel) embryos.
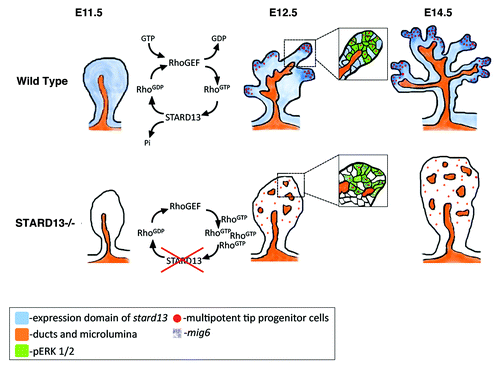
Stard13 is expressed in the developing pancreatic epithelium starting at E10.5. Around the onset of branching morphogenesis, Stard13 becomes enriched at the tip domains of the epithelium, where the multipotent pancreatic progenitors reside.Citation5,Citation10Pdx1-Cre-mediated deletion of Stard13 in the developing pancreas [hereafter referred to as Stard13PA-deleted] resulted in defective branching morphogenesis, disorganized “tip and trunk” domain tissue-architecture and subsequent postnatal organ hypoplasia.Citation16 Importantly, the same phenotype was observed upon the deletion of Stard13 gene expression ubiquitously using a CMV-Cre-deleter mouse strain, indicating that Stard13 acts as a pancreas tissue-specific GTPase regulator.Citation16 The analysis of the Stard13 mutant phenotype emphasized the tight connection between growth and morphogenesis in the developing pancreas (). We therefore set out to establish how the two processes are integrated and the role of the Rho signaling in this context.
Since pancreas organ size relies on the expansion of the progenitor cell reservoir in the embryo,Citation9 we analyzed if the loss of Stard13 affects the number and survival of pancreatic progenitor cells (Pdx1+;Cpa1+ cells). No significant difference in cell death was detected between wild-type (WT) and mutant littermates, whereas the number of proliferating cells was strongly reduced in Stard13PA-deleted embryos at E12.5. Strikingly, the proliferative defects almost exclusively affected the progenitor cells (Cpa1+;pHH3+ cells), while proliferation rate in the rest of the epithelium was similar to control counterparts.Citation16 In addition to a decreased proliferative activity, Cpa1+ progenitor cells appeared mislocalized and randomly distributed throughout the tissue, being the tip structures not properly formed in the mutant.Citation16 These observations suggest that the RhoGAP Stard13 plays an important role in sustaining proliferating progenitor cells in the embryonic pancreas.
Next, we sought to test the hypothesis whether the defects in proliferation were the consequences of perturbed tip morphogenesis or at the origin of it. Our detailed phenotypic analysis suggests that loss of Stard13 affects pancreatic epithelium starting from E11.5, which coincides with the initiation of the transition from a stratified unpolarized epithelium into a polarized monolayer of cells.Citation16,Citation19 Upon closer examination of cell morphology, polarity, and cytoskeletal organization, we observed that Stard13PA‑deleted embryos show major problems in epithelial remodeling. Specifically, Stard13 mutant cells remained stratified, cuboidal in shape and did not properly form and resolve “rosette-like” structures.Citation16,Citation20 In addition, we found that mutant cells display alterations in the actomyosin cytoskeletal organization, including not only an accumulation of F-actin and myosin, but also their irregular distribution throughout the cytoplasm.Citation16 Collectively, these results suggest that Stard13 is required for ensuring epithelial remodeling and initiation of branching in the pancreas. The fact that epithelial defects (e.g., aberrant cell shape and F-actin accumulation) were the earliest detectable defects in the mutant pancreas, while reduction in cell proliferation became obvious starting at E12.5, suggests a temporal succession of events, in which morphogenetic defects would underlay and, possibly, cause defective proliferation ().
As Stard13 contains a conserved RhoGAP domain, we anticipated it being able to regulate Rho in the developing pancreas. Accordingly, using pull down and immunolocalization assays, we demonstrated that mutant tissues accumulate elevated levels of active (GTP-bound) Rho, indicating that Stard13 is required to spatio-temporally limit Rho activity in the pancreas.Citation16 To determine whether the observed phenotype was due to uninhibited Rho activity, we used an ex vivo pancreatic culture system and performed rescue experiments using pharmacological inhibitors of Rho in Stard13 mutant pancreas.Citation5,Citation21 Importantly, inhibition of Rho in Stard13PA-deleted explants partially rescued branching and proliferation defects and restored asymmetric F-actin distribution in epithelial cells.Citation16 These findings indicate that Stard13 controls epithelial remodeling and morphogenesis by regulating Rho/actin signaling axis within the pancreatic epithelium. In addition, these results unveiled a novel role for actin cytoskeletal dynamics at the onset of pancreatic branching morphogenesis. The next question was whether the same Rho-actin signaling cascade may directly influence proliferation in the pancreatic epithelium and how.
Actin, which is best known as a cytoskeletal component, also participates in the control of gene expression, influencing the Serum Response Factor (SRF) transcriptional cascade.Citation22 Accordingly, in parallel with the accumulation of F-actin in the Stard13PA-deleted epithelium, we reported an induction of the SRF-dependent transcriptional activity and of its transcriptional co-activator the Megakaryoblastic leukemia-1 (MAL).Citation16 In particular, among others, we found elevated levels of the MAL/SRF downstream targets Mig6 and Zfp36, which are anti-proliferative signals and known to negatively regulate the Mitogen-Activated Protein Kinase (MAPK) signaling.Citation16,Citation23-Citation25 This finding is particularly intriguing, as we measured a concomitant reduction of the MAPK/pERK signaling activity in the Stard13 mutant as compared with WT pancreas.Citation16 Furthermore, decreased levels of pERK1/2 in Stard13 mutants were rescued to near WT levels by specific inhibition of Rho.Citation16 Collectively, these findings suggest the convergence of the Rho-actin signaling into the MAPK cascade through modulation of SRF targets, providing a mechanistic explanation for the reduced proliferation in Stard13PA-deleted pancreatic epithelium.
In line with this, the selective inhibition of downstream Rho/SRF transcriptional signaling rescued the Stard13 mutant cell proliferation defects.Citation16 Nevertheless, upstream inhibition of the Rho signaling using the C3 exotoxin molecule results in a more effective rescue of the Stard13PA-deleted defects than the transcriptional disruption of the SRF pathway using the compound CCG1423.Citation16 These observations reinforce the notion that cell shape and cytoskeleton are at the origin of the phenotypic defects in the Stard13 mutant, being not completely eliminated by a downstream rescue at the transcriptional level. Also, these results suggest possible mechanical effects of cell shape and actomyosin cytoskeleton on cell division in the developing pancreas that might be independent of the SRF/MAPK pathway. For example, the process of cytokinesis requires the formation and ingression of a cleavage furrow generated by the constriction of an actomyosin contractile ring anchored to the cell membrane by cytoskeletal proteins.Citation26 The cytokinetic apparatus is initiated by the activation of Rho in a precisely defined “zone” of the cell membrane and relies on tightly regulated cycling between GTP/GDP-bound states.Citation27 Thus, it is conceivable that the RhoGAP activity of Stard13 might be necessary during cytokinesis and the accumulation of active Rho antagonizes proliferation by impairing successful completion of cytokinesis.Citation28 Similarly, Rho being a central regulator of the cytoskeleton might also be involved in the orientation of cell divisions.Citation27,Citation29 For instance, a role for RhoA in the maintenance of spindle orientation has been shown in the chick neuroepithelium, controlling thereby the balance between planar and apico-basal divisions.Citation30 Oriented cell division might indeed be a mechanism (1)to expand the progenitor pool and (2) to generate diversity along the epithelial branches in the developing pancreas too. Stard13 might act a tissue-specific regulator of Rho cycling to ensure correct spindle orientation in a critical time-window of pancreas development. Alternatively, elevated Rho levels might affect proliferation of pancreatic progenitors through its mechanical control on cell-shape. Observations in other developing epithelia suggested that changes in cytoskeletal tension mediated by Rho signaling through ROCK might also establish spatial differentials in cell growth.Citation2,Citation31,Citation32 Further analyses will help to elucidate possible different and redundant functions of Rho and its regulator Stard13 in pancreatic growth and morphogenesis.
In several tissues and organ systems, it has been shown that stem cells reside in specific “niches” or microenvironments.Citation10,Citation33,Citation34 Similarly, the multipotent progenitors of the embryonic pancreas once properly allocated to the tips of the branches may define a supporting “niche.” For instance, mesenchymal cells surrounding the branching tips may signal to the epithelium, maintaining the progenitor state of the tip cells, while the cells more distant from the mesenchyme lose that influence and become restricted in their fate. Consistently, our study unveiled a differential distribution of active pERK signaling in the emerging tips, suggesting a crosstalk between epithelial progenitor tip cells and its surrounding mesenchyme (FGF and EGF source) that possibly sustains their proliferation and expansion.Citation16
In conclusion, based on our findings, we propose Stard13 as an intrinsic regulator of the number of pancreatic progenitors through the establishment of the pancreatic tip domain. From a mechanistic point of view, our data define a reciprocal interaction between the actin-MAL/SRF signaling and the MAPK signaling to locally regulate progenitor cells proliferation in the pancreas (). As such, the initial progenitor pool can expand and reach proper final size, which predetermines pancreas organ size in the adult.Citation9,Citation10 In the long run, full comprehension of the relationship between pancreatic progenitor cells and tissue-architecture will aid in defining new and more efficient strategies for expansion of pancreatic progenitors and generation of insulin producing β-cells from stem cells or progenitor cells as a potential cure to diabetes.
| Abbreviations: | ||
| E | = | embryonic stage |
| EGF | = | epidermal growth factor |
| FGF | = | fibroblast growth factor |
| GAP | = | GTPases-activating protein |
| MAL | = | megakaryoblastic leukemia-1 |
| MAPK | = | mitogen-activated protein kinase |
| SRF | = | serum response factor |
| WT | = | wild type |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Research in the Spagnoli lab. is funded by the Helmholtz Association, FP7-IRG-2008-ENDOPANC grant and ERC-2009-Starting HEPATOPANCREATIC Grant.
References
- Hogan BLM, Kolodziej PA. Organogenesis: molecular mechanisms of tubulogenesis. Nat Rev Genet 2002; 3:513 - 23; http://dx.doi.org/10.1038/nrg840; PMID: 12094229
- Nelson C.. Geometric control of tissue morphogenesis. Biochim Biophys Acta 2009; 1793:903 - 10
- Michael L, Davies JA. Pattern and regulation of cell proliferation during murine ureteric bud development. J Anat 2004; 204:241 - 55; http://dx.doi.org/10.1111/j.0021-8782.2004.00285.x; PMID: 15061751
- Nogawa H, Morita K, Cardoso WV. Bud formation precedes the appearance of differential cell proliferation during branching morphogenesis of mouse lung epithelium in vitro. Dev Dyn 1998; 213:228 - 35; http://dx.doi.org/10.1002/(SICI)1097-0177(199810)213:2<228::AID-AJA8>3.0.CO;2-I; PMID: 9786423
- Horb LD, Slack JM. Role of cell division in branching morphogenesis and differentiation of the embryonic pancreas. Int J Dev Biol 2000; 44:791 - 6; PMID: 11128573
- Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn 2011; 240:530 - 65; http://dx.doi.org/10.1002/dvdy.22584; PMID: 21337462
- Puri S, Hebrok M. Cellular plasticity within the pancreas--lessons learned from development. Dev Cell 2010; 18:342 - 56; http://dx.doi.org/10.1016/j.devcel.2010.02.005; PMID: 20230744
- Spagnoli FM. From endoderm to pancreas: a multistep journey. Cell Mol Life Sci 2007; 64:2378 - 90; http://dx.doi.org/10.1007/s00018-007-7184-x; PMID: 17565443
- Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature 2007; 445:886 - 91; http://dx.doi.org/10.1038/nature05537; PMID: 17259975
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell 2007; 13:103 - 14; http://dx.doi.org/10.1016/j.devcel.2007.06.001; PMID: 17609113
- Villasenor A, Chong DC, Henkemeyer M, Cleaver O. Epithelial dynamics of pancreatic branching morphogenesis. Development 2010; 137:4295 - 305; http://dx.doi.org/10.1242/dev.052993; PMID: 21098570
- Van Aelst L, Symons M. Role of Rho family GTPases in epithelial morphogenesis. Genes Dev 2002; 16:1032 - 54; http://dx.doi.org/10.1101/gad.978802; PMID: 12000787
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature 2002; 420:629 - 35; http://dx.doi.org/10.1038/nature01148; PMID: 12478284
- Greiner TU, Kesavan G, Ståhlberg A, Semb H. Rac1 regulates pancreatic islet morphogenesis. BMC Dev Biol 2009; 9:2; http://dx.doi.org/10.1186/1471-213X-9-2; PMID: 19126199
- Kesavan G, Sand FW, Greiner TU, Johansson JK, Kobberup S, Wu X, et al. Cdc42-mediated tubulogenesis controls cell specification. Cell 2009; 139:791 - 801; http://dx.doi.org/10.1016/j.cell.2009.08.049; PMID: 19914171
- Petzold KM, Naumann H, Spagnoli FM. Rho signalling restriction by the RhoGAP Stard13 integrates growth and morphogenesis in the pancreas. Development 2013; 140:126 - 35; http://dx.doi.org/10.1242/dev.082701; PMID: 23175628
- Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell 2007; 99:67 - 86; http://dx.doi.org/10.1042/BC20060086; PMID: 17222083
- Sordella R, Jiang W, Chen G-C, Curto M, Settleman J. Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell 2003; 113:147 - 58; http://dx.doi.org/10.1016/S0092-8674(03)00271-X; PMID: 12705864
- Hick A-C, van Eyll JM, Cordi S, Forez C, Passante L, Kohara H, et al. Mechanism of primitive duct formation in the pancreas and submandibular glands: a role for SDF-1. BMC Dev Biol 2009; 9:66; http://dx.doi.org/10.1186/1471-213X-9-66; PMID: 19126199
- Zallen JA, Blankenship JT. Multicellular dynamics during epithelial elongation. Semin Cell Dev Biol 2008; 19:263 - 70; http://dx.doi.org/10.1016/j.semcdb.2008.01.005; PMID: 18343171
- Petzold KM, Spagnoli FM. A system for ex vivo culturing of embryonic pancreas. J Vis Exp 2012; 66:e3979; PMID: 22951988
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007; 316:1749 - 52; http://dx.doi.org/10.1126/science.1141084; PMID: 17588931
- Descot A, Hoffmann R, Shaposhnikov D, Reschke M, Ullrich A, Posern G. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol Cell 2009; 35:291 - 304; http://dx.doi.org/10.1016/j.molcel.2009.07.015; PMID: 19683494
- Ferby I, Reschke M, Kudlacek O, Knyazev P, Pantè G, Amann K, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med 2006; 12:568 - 73; http://dx.doi.org/10.1038/nm1401; PMID: 16648858
- Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 2006; 16:588 - 96; http://dx.doi.org/10.1016/j.tcb.2006.09.008; PMID: 17035020
- Glotzer M. Animal cell cytokinesis. Annu Rev Cell Dev Biol 2001; 17:351 - 86; http://dx.doi.org/10.1146/annurev.cellbio.17.1.351; PMID: 11687493
- Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol 2005; 15:651 - 8; http://dx.doi.org/10.1016/j.tcb.2005.10.006; PMID: 16243528
- Morin P, Flors C, Olson MF. Constitutively active RhoA inhibits proliferation by retarding G(1) to S phase cell cycle progression and impairing cytokinesis. Eur J Cell Biol 2009; 88:495 - 507; http://dx.doi.org/10.1016/j.ejcb.2009.04.005; PMID: 19515453
- Morin X, Bellaïche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell 2011; 21:102 - 19; http://dx.doi.org/10.1016/j.devcel.2011.06.012; PMID: 21763612
- Roszko I, Afonso C, Henrique D, Mathis L. Key role played by RhoA in the balance between planar and apico-basal cell divisions in the chick neuroepithelium. Dev Biol 2006; 298:212 - 24; http://dx.doi.org/10.1016/j.ydbio.2006.06.031; PMID: 16860308
- Moore KA, Polte T, Huang S, Shi B, Alsberg E, Sunday ME, et al. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev Dyn 2005; 232:268 - 81; http://dx.doi.org/10.1002/dvdy.20237; PMID: 15614768
- Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, et al. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A 2005; 102:11594 - 9; http://dx.doi.org/10.1073/pnas.0502575102; PMID: 16049098
- Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell 2004; 116:769 - 78; http://dx.doi.org/10.1016/S0092-8674(04)00255-7; PMID: 15035980
- Rawlins EL, Clark CP, Xue Y, Hogan BLM. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 2009; 136:3741 - 5; http://dx.doi.org/10.1242/dev.037317; PMID: 19855016