Abstract
Current literature using biochemical assays, structural analyses and genetic manipulations has reported that the key factors associated with the faithful matching of the initiator met-tRNA to the start codon AUG are eIF1, eIF1A and eIF5. However, these findings were in each case based upon the utilization of a single mRNA, perhaps with variations. In an effort to evaluate this general finding, we tested six different mRNAs. Our results confirm that these three proteins are important for start site selection. However, two additional findings would not have been predicted. The first is that eIF1 plays a major role in selecting against start codons that are in close proximity to the 5′ end of the mRNA (i.e., less than 21 nucleotides). Second, the addition of eIF5B had nearly the same affect as the addition of eIF5. This is unexpected given the different roles that eIF5 and eIF5B have been proposed to play in the 80S initiation pathway. Finally, although many of the mRNAs appear to respond qualitatively in a similar manner, the quantitative differences noted suggest that there is still some mRNA specific character to our findings. This character may be the length of the 5′ UTR, involvement of an IRES element, secondary structure either 5′ or 3′ of the start codon or specific sequence/structure elements that interact with RNA binding proteins or the ribosome.
Introduction
Early insights into start site selection came from the laboratories of Dr. Marilyn Kozak and Dr. Thomas Donahue who used cell free protein synthesis and genetics, respectively, to determine elements important for authentic start site recognition. Dr. Kozak’s work defined the nucleotide sequences that were most favorable for authentic AUG recognition and also evaluated the influence of downstream secondary structure.Citation1-Citation3 Dr. Donahue determined by genetic mutations the proteins associated with start site recognition and defined 5 SUI (suppressor of initiation) mutations: SUI1 (eIF1), SUI2 (eIF2α), SUI3 (eIF2β), SUI4 (eIF2γ) and SUI5 (eIF5).Citation4-Citation6 More recent studies from a number of laboratories have added detail in the kinetic and genetic interactions of translation factors and start site selection and the physical location of these factors on the surface of the 40S subunit.Citation7-Citation24 Much of this information is captured nicely in of a review by Hinnebusch.Citation25 In brief, in the early steps of initiation, the binding of eIF1 and eIF1A appears to cause a conformational change in the 40S subunit that places it in an “open” conformation that now can accommodate the binding and placement of the mRNA on the 40S subunit. Scanning of the mRNA occurs prior to or following the hydrolysis of GTP in the ternary complex (may depend on the length of the 5′ UTR) but in the absence of the release of the Pi from eIF2, the complex remains stable. However, the correct matching of the initiator tRNA with the AUG start codon triggers release of eIF1 and the subsequent release of Pi allowing for the conversion of this complex to the “closed” conformation (for greater detail, see reviews 25–28). What is currently uncertain is how valid this general pathway is in comparative studies between yeast (with its multifactor complex or MFC)Citation29 and the mammalian system where either individual factors or perhaps pairs of factors might be interacting with one another.
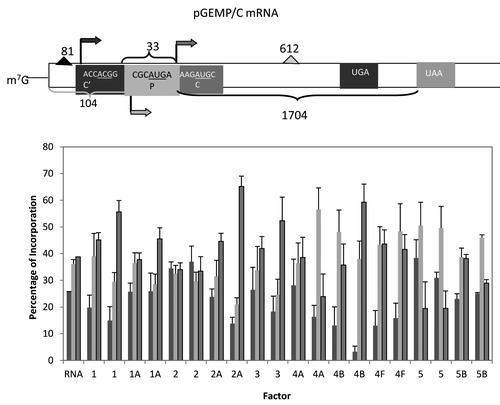
Figure 5. Influence of added initiation factors on the translation of the pGEMP/C mRNA. Above the bar graph is a representation of the pGEMP/C mRNA. The bar graph shows the relative levels of the long, medium and short forms of the reporter protein observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor. C’ is an in frame extension of C while P is expressed from a reading frame different from C’ and C.
Much of the above studies have utilized a single mRNA base transcript which was then mutated to determine the importance of: sequence context around the initiating AUG codon; secondary structure either 5′ or 3′ of the initiating AUG codon; influence of specific initiation factors (or mutants thereof). In this study, we have examined six different mRNAs for the influence of initiation factors on start site selection. These studies have confirmed the importance of eIF1, eIF1A and eIF5 in start codon selection. Additionally, we have found that eIF5B appears to have a similar influence as eIF5 even though previous studies have shown it to function after the start site has been selected (see model pathways in the following reviews: ref. Citation25–Citation30). We also found that eIF1 has a very dramatic influence on start site utilization when the initiating AUG codon is close to the 5′ m7G cap. And while the influence of factors on start site selection was qualitatively similar in many instances, the quantitative behavior was somewhat different. These differences may reflect differences in 5′ UTR length, secondary structure around the start site (either 5′ or 3′; note that this might also be influenced by the binding of proteins to this region as well), or the three dimensional shape of the mRNA (as an mRNP). The important feature is that the characterization of the influence of specific translation initiation factors on start site selection continues to have an mRNA specific component.
Results
As a starting point to determine if translation factor activity could alter start site selection, we chose to use nuclease-treated, rabbit reticulocyte lysate. The major reason for choosing this system is its known synthetic rate, essentially equal to the in vivo rate of protein synthesis. We anticipated that the shifting from one start site to another would be a sensitive transition and thus, felt that only the most active system might reveal any differences. Second, by using this system under conditions when mRNA was limiting, we anticipated that we would be at the most sensitive position to look for differences, within the linear range. In particular, we chose to not use conditions of saturating mRNA or mRNA competition which would complicate interpretation. Presented in are the results of our titration of the different mRNAs into the assay system and the optimal time point determination (middle to end of the linear increase in hot TCA precipitable radioactive methionine). Although there was some variation, the optimal concentration of mRNA was about 0.4 μg per 25 μl reaction with an optimal incubation time of 40–50 min.
Figure 1. Summary of the optimal mRNA concentrations and reaction times. As described in results, optimal mRNA levels and reaction times for in vitro translation were determined independently and are shown in the figure. Determination of the optimal conditions was based upon hot TCA precipatable [35S]methionine with aliquots taken for the utilization of 0.2, 0.4, 0.6, 0.8 and 1.0 μg of added mRNA or at 0, 10, 20, 30, 40, 60 and 80 min. when an optimal amount of mRNA had been determined.
![Figure 1. Summary of the optimal mRNA concentrations and reaction times. As described in results, optimal mRNA levels and reaction times for in vitro translation were determined independently and are shown in the figure. Determination of the optimal conditions was based upon hot TCA precipatable [35S]methionine with aliquots taken for the utilization of 0.2, 0.4, 0.6, 0.8 and 1.0 μg of added mRNA or at 0, 10, 20, 30, 40, 60 and 80 min. when an optimal amount of mRNA had been determined.](/cms/asset/b59bacc9-b524-427e-8e0b-4bf6c902dd95/ktrs_a_10924419_f0002.gif)
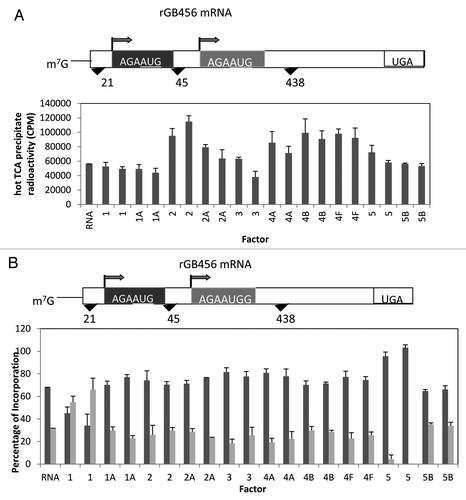
We began our investigation with the synthetic construct of a rabbit β globin mRNA with two identical AUG start sites (AGAAUGG) placed 45 nucleotides apart (a gift from Dr. Stan Tahara).Citation31 This start site conforms to the standard optimal sequence predicted by Kozak with a purine at -3 and a G at +4 (the A in AUG being nucleotide +1).Citation1 Thus, there should be no favoritism due to the start site context. As can be seen in , the addition of a number of initiation factors failed to influence total incorporation into TCA precipitable radioactivity. The general exceptions were the proteins associated with binding the initiator tRNA to the 40S subunit (eIF2) and those associated with activation and binding of the mRNA to the 43S complex (eIF4A, eIF4B, eIF4F) which stimulated protein synthesis about 2-fold. However, a much different outcome was noted when one determined the amount of protein made from either the first or second start site (). While many additions did not alter the roughly 70%/30% ratio of first site to second site starts, two additions were quite different. The addition of eIF1 led to the preferred utilization of the second start site while the addition of eIF5 lead to the almost exclusive use of the first start site.
Figure 2. Influence of added initiation factors on the translation of the rGB456 mRNA. Protein synthesis was performed as described in Methods. Panel A shows the total hot TCA precipitable radioactivity obtained in the presence of no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition). For simplicity, the eIF designation is not included in front of the number for each factor. Panel B shows the relative amount of the long (initiated at the first AUG) and the short (initiated at the second AUG) form of the reporter protein (% of the 2 forms such that the long form + the short form = 100%). The relative amount of each protein was determined as described by Laemmli,Citation32 followed by resolution of the two protein bands by SDS PAGE. Dried gels were exposed to X-ray film and then quantitation of the bands was performed by use of a phosphoImager followed by analysis using Imagequant. Above the bar graph is a cartoon representation of the m7G capped mRNA used.
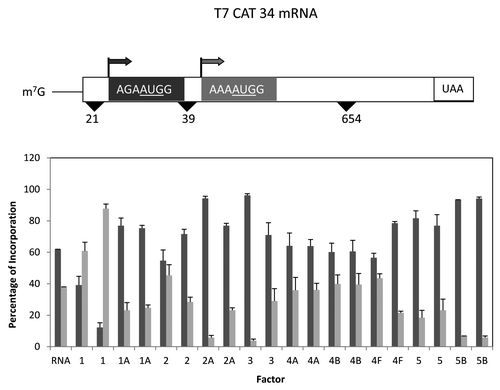
A second related mRNA was the T7CAT34 mRNA (also provided by Dr. Stan Tahara) which was slightly different in that there was a different reporter protein and the 5′ UTR was slightly shorter.Citation31 In contrast to the β globin mRNA, little stimulation of translation was observed with the eIF4 group of proteins, but a similar 2-fold increase was noted with added eIF2 (data not shown). Although there appeared to be little change in total synthesis, the addition of different translation factors did cause a dramatic change in the distribution of start site selection (). The addition of eIF1A, eIF2A, eIF3, eIF5 and eIF5B all increased the proportion of first start site utilization changing the distribution from roughly 60%/40% (first/second start site) to as much as 90%/10%. In contrast, the addition of eIF1 lead to the preferred utilization of the second start site with a ratio of roughly 30%/70% depending on the amount of eIF1 added as was also noted with the β globin mRNA.
Figure 3. Influence of added initiation factors on the translation of the T7CAT34 mRNA. At the top of the figure is a representation of the T7CAT34 mRNA and below is the relative amount of the long and short form of the reporter protein made in the presence of added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor.
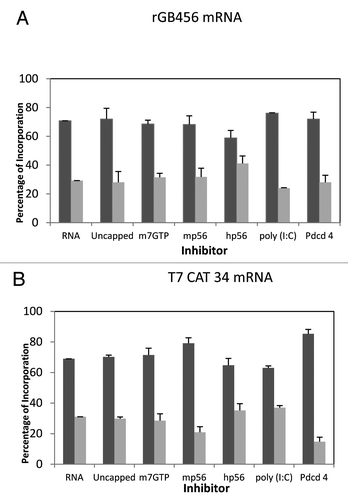
Given the similarity of the mRNAs, but different results obtained when adding the additional translation factors, we wondered if similar changes might be observed when initiation factor concentrations were reduced. Since intact reticulocyte lysate was being used, the primary way to effectively lower initiation factor concentrations was the use of inhibitors (m7GTP, mouse p56,Citation33 human p56,Citation34 poly(I:C) or Pdcd4Citation35). Based upon previous studies, these inhibitors would be expected to reduce the levels of active/effective eIF4F (m7GTP, mp56), eIF2 (poly(I:C), hp56) or eIF4A (Pdcd4). All of the inhibitors reduced expression by 40% to 70% except m7GTP which was ineffective. The outcome indicated essentially no influence on start site selection except for a modest affect of Pdcd4 on the T7CAT34 mRNA ().
Figure 4. Influence of protein synthesis inhibitors on expression from the rGB456 and T7CAT34 mRNAs. Panel A – Protein synthesis was performed using the rGB456 mRNA as described in with the addition of protein synthesis inhibitors which included: 100 μM m7GTP, 122 nM mp56, 180 nM hp56, 600 pg of poly(I:C) or 0.6 μg of Pdcd4. For each inhibitor, the reaction mixture was incubated with the inhibitor for 15 min. at 30°C prior to the start of the reaction by the addition of mRNA. The control reaction (RNA) was also pre-incubated followed by the addition of the mRNA. Levels of inhibition observed ranged from 40 to 70% except for m7GTP where little inhibition was observed. Shown are the relative amounts of the long form and short forms of the protein made. Panel B – The same analysis as in panel A was performed with the T7CAT34 mRNA.
An alternative view of start site selection is how important is the context for initiation, both the start codon and the nucleotide context preceding the start codon. The next two mRNAs examined were pGEMP/C mRNA derived from the viral RNA from Sendai virusCitation36 and the Pim2 mRNA, the mRNA for an oncogenic serine/threonine kinase.Citation31,Citation37 In the case of pGEMP/C, there are three possible start sites: the first in good context but with a ACG start codon; the second in poor context with an AUG start codon and the third in good context with an AUG start codon (see ). Four of the translation factors (eIF1, eIF2A, eIF3 and eIF4B) shifted the start site selection from about equal for all three to a preferred use of the third start site, the only one with a strong context and AUG start codon. At the higher level of added factor, there was some increase in the level of expression from the second start site with added eIF4A, eIF5 and eIF5B but these shifts were not as pronounced.
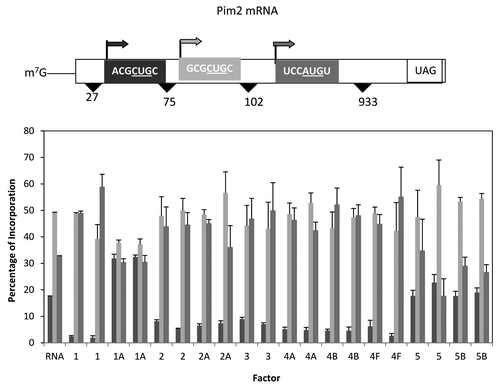
For the Pim2 mRNA, again, three start sites were possible, the first two in good context but with CUG as the start codon with the third start site in poor context but with an AUG start codon (). Similar to the β globin and T7CAT34 mRNAs, the first start site was relatively close to the 5′ end of the mRNA. And as was noted with those mRNAs, the addition of eIF1 shifted start site selection dramatically away from the first start site while the addition of eIF1A doubled the expression from the first start site. Most of the other factor additions appeared to reduce slightly initiation at the first start site with little change in the ratio of the use of the latter two sites. However, the addition of either eIF5 or eIF5B lead to the preferred utilization of the second start codon at the expense of the third.
Figure 6. Influence of added initiation factors on the translation of the Pim2 mRNA. Above the bar graph is a representation of the Pim2 mRNA. The bar graph shows the relative levels of the long, medium and short forms of the reporter protein observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor.
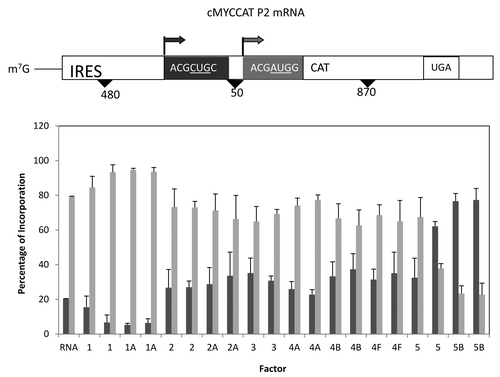
A much different mRNA tested was the cMYCCATP2 mRNA which had a considerably longer 5′ UTR and contained the cMyc IRES element (see ).Citation38 The use of the natural mRNA sequence does alter start site selection a bit in that the first start site utilizes a CUG start codon (in good context) in contrast to the AUG codon used for the second start site. Addition of translation initiation factors failed to stimulate translation (data not shown), but did alter the utilization of start sites. The addition of either eIF1 or eIF1A reduced the utilization of the first start site while the addition of either eIF5 or eIF5B lead to the preferred utilization of the first start site containing CUG as the initiation codon. These latter results are similar to those observed above for eIF5 and eIF5B and suggest that the process of initiation (cap-dependent and IRES-mediated) is sensitive to the level of these proteins.
Figure 7. Influence of added initiation factors on the translation of the cMYCCATP2 mRNA. Above the bar graph is a representation of the cMYCCATP2 mRNA. The bar graph depicts the relative levels of the long and short forms of the reporter protein observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor.
Our final mRNA was based upon the mRNA that encodes eIF4G1 which has the potential of 4 start sites, one 5′ of the putative IRES element and three downstream of this element.Citation39 The first, third and fourth start sites are all in good context with AUG start codons while the second start site is in poor context. The in frame AUG between the third and fourth start sites (which would be in a poor context) did not yield a detectable product and thus, it is assumed that this AUG does not serve any initiation function. The addition of translation factors failed to stimulate translation although addition of eIF5B did show significant inhibition of overall translation (about 50%; data not shown). With respect to start site selection, most of the factors had little influence although there was a modest increase for start site three with added eIF1. The addition of eIF5B was the only factor that led to the preferred utilization of the second start site, in part as a quantitative reduction in the utilization of start site one.
Discussion
Of the many translation factors associated with start codon selection, eIF1, eIF1A and eIF5 have been the most studied. In general terms, eIF1 and eIF1A have been associated with increased fidelity of recognition of the start codon and elevated eIF5 activity has been associated with decreased fidelity of AUG recognition. These interpretations have been identified through genetic screens in yeast, the use of molecular genetics in mammalian cells and emerging structural studies that have utilized either cryo-EM or high field NMR. An extremely thorough review of this topic has recently been published.Citation25 In this report, five of the six mRNAs studied were influenced by changes in these three proteins with only the eIF4G mRNA showing no shift in start codon selection with increases in these factors.Unexpectedly, unlike previous studies, we also found that eIF5B had an affect similar to that of eIF5 for some mRNAs. This finding may reflect the observation that in model mammalian systems, eIF5B is capable of triggering the hydrolysis of GTP in the ternary complex (although perhaps not as efficiently as eIF5).Citation40-Citation42
eIF1
Although associated with high fidelity recognition of the AUG codon,Citation13,Citation18,Citation23,Citation25,Citation43 in the three mRNAs with short 5′ UTRs (18–21 nucleotides), increased eIF1 caused a dramatic reduction in the utilization of the first start codon, even though the context around the start codons was strong. Recently, a unique element has been defined that enhances translation from start codons near the 5′ end referred to as the TISU element (translation initiator of short 5′ UTR) that has the identified sequence SAASAUGGCGGC where S can be either C or G.Citation54 Shown below is a comparison of this sequence with the first start sites found in the 3 mRNAs with short 5′ UTRs.
mRNA START SITE
TISU SAAS-AUG-GCGGC
rGB456 GAGA-AUG-GUGAG
T7CAT34 CAGA-AUG-GUAAG
Pim2 UGGG-CUG-GCGCG
In a direct comparison of mRNAs with an 11 nucleotide 5′ UTR, the addition of eIF1 to the extract did not alter start site selection for the TISU element as the first start site, but did favor utilization of the second start site in an non-TISU mRNA.Citation44 Given that none of the upstream start sites in our test mRNAs are a good match to the TISU element, our results are consistent with those published. In the two instances where the AUG is in a poor or good context, not surprisingly, added eIF1 favored the downstream AUG in good context although even in the untreated RRL, the downstream AUG was already 40% (pGEMP/C) or 80% (cMYCCATP2) of the initiation to begin with.
eIF1A
As anticipated, addition of eIF1A tended to enhance initiation at AUG codons in strong context. This was noted most strongly in the cMYCCATP2 mRNA where initiation at the upstream CUG start codon was reduced from 20% to about 7% (). A similar, but less pronounced shift to a better start codon context was also seen with the pGEMP/C mRNA (). It is possible that the reduced shift is in part a reflection of two upstream start signals in poor context and thus the affect was muted.
eIF5
Based upon the proposed function of eIF5 (triggers the hydrolysis of the GTP in the ternary complex of eIF2•GTP•Met-tRNAi), it is not surprising that the addition of eIF5 might enhance the hydrolysis of the GTP in the ternary complex thereby favoring start sites positioned more to the 5′ end of the UTR.Citation45 Indeed, this was an early observation for mutations in eIF5 with enhanced activity in activating the hydrolysis of GTP in the ternary complex resulting in a Sui phenotype.Citation6 The physical interpretation has been that the hydrolysis of GTP in the ternary complex relaxes the specificity of the “ternary complex” for the matching of the initiator met-tRNA with a start codon. Our results demonstrated that this “relaxed” specificity also plays out positionally in that even when there is no difference in the start codon context, the 5′ start codon is preferred when excess eIF5 is present.
eIF5B
Current 80S pathways have eIF5 triggering the hydrolysis of the GTP in the ternary complex and the release of eIF2.Citation25-Citation28,Citation30,Citation46 Subsequently, a second GTP and eIF5B are required to accomplish subunit joining. However, model studies have shown that eIF5B can trigger the hydrolysis of GTP bound to eIF2, either as monitored in 43S complex formation or as methionyl-puromycin synthesis.Citation40,Citation41 Consistent with these older observations, we found that the addition of eIF5B appeared to have affects highly similar, but not identical, to those seen with additional eIF5. We anticipate that these findings may also reflect an enhanced hydrolysis of the GTP in the ternary complex as was seen in model systems.
A concern expressed during review was that the eIF5B preparation might have been contaminated with eIF5 and that it was this eIF5 causing the affects attributed to eIF5B. A direct test of the eIF5B preparation by western blot indicated that it contained very low levels of eIF5, in the 1 to 3% range (data not shown) and thus would not have been enough to even yield the affects seen with added eIF5. Second, the results obtained in where added eIF5 led to the exclusive use of the first start site are dissimilar to those for added eIF5B which was essentially unchanged from the control. In contrast, in and , the shift to the utilization of the first start site is much more pronounced with added eIF5B than eIF5. This is inconsistent with the same molecule being responsible for the observed change in start codon selection for these three mRNAs and thus the affects observed are attributed solely to eIF5B.
Other factors
Two other factors also had some influence on start site selection. For the T7CAT34 mRNA (), both added eIF2A and eIF3 enhanced expression from the 5′ start codon even though both were in good context. In contrast, both added eIF2A and eIF3 favored the use of the 3′ most start codon in the pGEMP/C mRNA, the start codon in the best context (i.e., not ACCACGG or CGCAUGA). At this point in time, there is no simple explanation for why the different preferences (5′ vs. 3′) and one assumes that this might be more of an mRNA specific affect rather than a generalizable characteristic of either factor.
Figure 8. Influence of added initiation factors on the translation of the eIF4G1 mRNA. Above the bar graph is a representation of the eIF4G mRNA. The bar graph shows the relative levels of the four forms of the reporter protein (as indicated by the arrows) observed in the presence of the added initiation factors. The result of having no added initiation factors (RNA) or 1X or 2X added initiation factor (the 1X value is the left most column for each factor addition) is shown. For simplicity, the eIF designation is not included in front of the number for each factor. No initiation was observed for the possible start codon UUGAUGA.
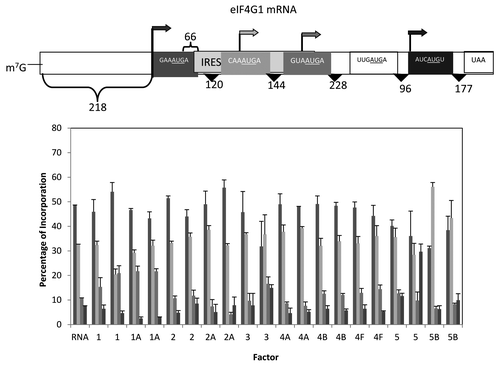
The eIF4G mRNA
The initial finding that the eIF4G mRNA contained an IRES element suggested that perhaps, as had been noted for IF3 or RF2 in the bacterial system, this provided a mechanism for autoregulation (i.e., low levels of eIF4F would favor IRES-mediated expression, high levels of eIF4F would favor cap-dependent translation thereby repressing IRES-mediated expression). Given the series of results of the first five mRNAs, it was surprising to find that the eIF4G mRNA appeared to be refractory to changes in levels of the initiation factors except for eIF5B which favored expression of the second start codon at the expense of the third and fourth start codons. It is not clear to us whether the lack of influence of added factors is in part a complication due to the low level of expression obtained with this mRNA (about one half to one third of most of the mRNAs examined).
The affects noted in this study of 6 different mRNAs are designed to be a mimic of possible changes in the level of initiation factor activity that may be the result of covalent modification or differential protein concentrations as a function of cellular development or the cell’s response to its environment. Of the proteins utilized in this study, eIF1, eIF2, eIF2A, eIF3 (multiple subunits), eIF4B, eIF4E, eIF4G, eIF5 and eIF5B are known to be phosphorylated and it is possible that other covalent modifications may occur for either these or other initiation factors as well.Citation47 Thus, the types of changes in start site utilization observed here could very readily be accomplished by cells. That said, it should be noted that the one surprise in our findings was that downregulation of translation factor activity did not appear to cause any significant change in start site utilization relative to untreated RRL. In part, this may reflect our initial assay conditions whereby we chose to deliberately use non-saturating levels of mRNAs to establish a more sensitive assay system. These conditions are unlikely to reflect what is occurring in other (in vivo) studies where cells are in log phase growth and the predominant translation occurring is that of the housekeeping proteins that are required for cell doubling and for whose mRNAs the translation is cap-dependent and efficient. However, these conditions may be much more relevant for whole animal studies where tissues respond to development, nutritional alterations or cellular stresses and are not in log phase growth. The other caveat is that we did not have useful inhibitors for those proteins that showed the most pronounced affects in start site selection (eIF1, eIF1A, eIF5, eIF5B) but rather for those proteins that are associated with the key regulatory points in the cap-dependent 80S pathway (eIF2 and eIF4F).
One concern in these studies was that the addition of an initiation factor might alter the balance of various complexes within the RRL. In this light, the following complexes have been reported to form, and in some instances, found to be quite stable: eIF3•ternary complex; eIF3•eIF4F; eIF4F•eIF4B; eIF1A•eIF5B. Thus, the addition of eIF3 might titrate either eIF2 (as the ternary complex) or eIF4F resulting in an effectively lower concentrations of free ternary complex or eIF4F (obviously, if these eIF3 complexes were part of the 80S pathway, then such a titration affect might not occur). In addition, some of the observed affects may reflect the disruption of the formation of the multifactor complex (MFC) as best studied in yeast.Citation26,Citation29 If the equivalent complex were to exist in mammalian systems, one could imagine a resulting imbalance (i.e., if the MFC was composed of eIF1, eIF2 (as the ternary complex), eIF3, and eIF5, then the addition of “extra” eIF3 might result in partial complexes of eIF3•eIF2•eIF5, eIF3•eIF2•eIF1 or eIF3•eIF1•eIF5 depending on the binding interactions between proteins thus reducing the concentration of the complete complex of all four proteins).
Is the RRL assay system a faithful reproduction of in vivo events? While in part the answer is still unknown, it was noted that for most of the mRNAs tested, the observed ratio of products in the absence of added factors was essentially the same as when these constructs were tested in tissue culture cells or as reported previously from using in vitro expression.Citation36,Citation38,Citation48 The major difference we noted between previous reports and our studies was for the mRNAs RGB456 and T7CAT34 where it had been observed that the addition of eIF4F increased the level of expression from the first start site.Citation31 In a series of control experiments using uncapped mRNAs, we were able to determine that this increase was most probably due to the presence of a higher proportion of uncapped mRNA used in the Tahara et al. study,Citation31 either as uncapped mRNA or an mRNA in which the cap was inverted such that the m7G residue was the first nucleotide in the regular RNA portion of the mRNA (our data; not shown). Thus, as a safe guard in the preparation of capped mRNAs, we feel that the use of the anti-reverse cap analog (ARCA) and a ratio of analog to GTP of at least 8 to 1 is required to insure that the results obtained primarily/only reflect the properties of a naturally capped mRNA even though this means that the yield of RNA from the transcription reaction will only be about one tenth that observed in the absence of an analog.
Our results are consistent with a growing body of literature that implicates eIF1, eIF1A and eIF5 as key determinants in start site selection, either as cap-dependent or IRES-mediated initiation. Additionally, as noted with the TISU mRNAs,Citation44 eIF1 also functions to discriminate against short 5′ UTRs when the sequence is not the TISU consensus (C/GAAC/GAUGGCGGC). In a manner that is currently not clear, eIF5B has properties similar to eIF5 which is surprising given their different roles in initiation (eIF5, as the GAP for the ternary complex and eIF5B as the GTP-dependent subunit joining factor). The interplay of eIF1, eIF1A and eIF5 in start site selection and the similar roles of eIF5 and eIF5B may reflect some evolutionary degeneracy that provides a protective affect against mutations in any one of the proteins. In this light, it is noted that others have reported on 80S complex formation with either only eIF5 or only eIF5B.Citation40,Citation41,Citation45,Citation49-Citation52 It is possible that a true kinetic comparison will be required to shed more light on the potential interchangeability of these two proteins, although eIF5 is an essential gene in yeast, but eIF5B is not (although the deletion of eIF5B results in a yeast strain with a slow growth phenotype).
While the above discussion sounds reasonable in the face of other existing data, the question is raised “But why should added protein, especially eIF1 or eIF1A, have any effect?” Based upon either purification or an examination of the 80S initiation pathways in most reviews, it would seem that these two proteins are bound early in the pathway and stably associated with the 40S subunit until their release, at some point after hydrolysis of GTP in the ternary complex. There are several possibilities. The first is that under conditions of either binding the mRNA or scanning, the association of these factors with the 40S subunit is represented by an equilibrium, generally favoring association but becoming closer to 100% associated in the presence of a higher concentration of factor. A second possibility would be that a higher concentration of factor might serve as “product inhibition” facilitating the “mis-binding” of the factor after the step in which it had just been released. A third possibility could be that the higher factor concentration is allowing a secondary binding, either to the ribosome or to another factor (in the case of eIF1A, binding to eIF5B might either activate or inactivate its function if the proteins formed a dimer in solution, which has been observed biochemicallyCitation53). The authors have no particular preference for these alternatives and clearly some other explanation may be correct. As is too often the case, further studies are required.
Finally, it is clear to us that not all mRNAs are the same and this is reflected in the different quantitative responses to the addition of initiation factors, especially eIF1, eIF1A, eIF5 and eIF5B. A simple example would be in , added eIF5 lead to almost 100% utilization of the first AUG (70% in control) while in , added eIF5 caused less of a change (from 60% to 80% utilization of the first AUG). In contrast, added eIF5B had no affect in but caused an even greater utilization of the first AUG in (from 60% to 95%). Thus, while it is likely that one can predict qualitatively a given response, the ability to predict quantitatively is not possible and changes in expression due to changes in the levels of initiation factor activity will continue to be specific to each mRNA.
Materials and Methods
Materials
Rabbit reticulocyte lysate (RRL) was obtained from Promega and was used as described in their technical manual titled “Rabbit reticulocyte lysate systems: Instructions for the use of products L4960 and 4151.” This lysate had been treated with micrococcal nuclease by Promega to reduce or eliminate endogenous globin mRNA. [35S]methionine was obtained from GE Health Sciences. Purified translation factors were purified from untreated rabbit reticulocyte lysate obtained from Green Hectares (Oregon, WI) as described previously.Citation46,Citation54-Citation60 The overall purifications using phosphocellulose (Whatman P11), DEAE cellulose (Whatman DE52), CM cellulose (Whatman CM52), sizing matrices (Sephadex G200, G150, G100; Pharmacia), and sucrose gradients in high salt led to the separation of the following factors, resolved from one another: eIF1A, eIF2, eIF2A, eIF3, eIF4A, eIF4B, eIF4F, eIF5A, and eIF5B. eIF1 and eIF5 were expressed in E. coli as His6 tagged proteins using plasmids kindly provided by Drs. Tatyana Pestova and Christopher Hellen (SUNY, Brooklyn) and purified as described by them. The inhibitors used in this study were: m7GTP purchased from Sigma, poly(I:C) from P-L Biochemicals, Pdcd4 was kindly provided by Drs. Hsin-Sheng Yang and Nancy H. Colburn (NIH), and human p56 and mouse p56 were kindly provided by Drs. Daniel J. Hui and Ganes Sen (Cleveland Clinic Foundation). Based upon concerns expressed by Drs. Dever and Lorsch, our eIF5B preparation was probed for contamination by eIF5 using a commercially available antibody (Santa Cruz). Using our bacterially expressed eIF5 as a control, the level of eIF5 in the eIF5B was estimated to be 1–3% by western blot.
Methods
Cell free translationCitation61,Citation62 – The standard reaction mixture (25 μl) contained: 17.5 μl of nuclease-treated RRL, 0.5 μl of a 19 amino acid mixture (minus methionine), 1 μl [35S]methionine (roughly 10 mCi/ml), 0.5 μl RNasin, 1 μl mRNA, and 4.5 μl of buffer (20 mM Tris•HCl, pH 7.5, 1 mM DTT, 0.1 mM EDTA, 100 mM KCl and 10% glycerol). The buffer is the same solution that the initiation factors are stored in. When initiation factors were added, the combined volume of initiation factors and buffer was 4.5 μl. For the 1X and 2X addition of initiation factors, 1X was equivalent to approximately 1 μg of factor and 2X was equivalent to 2 μg of factor except for eIF3 which was added at 5 and 10 μg due to its much greater molecular weight. In general, the 2X amount added would be roughly equivalent to doubling the concentration of that factor in the RRL (and more like 4–5 times the amount for the small proteins, eIF1 and eIF1A). These are also the amounts that generally were used in fractionated assay systems with purified components.Citation61 For each mRNA, a titration of RNA was performed monitoring the incorporation of [35S]methionine for 60 min. at 37°C. The level of each mRNA was selected that appeared to be about half saturating under these conditions and referred to as the optimum concentration in . Subsequently, at this mRNA level, a time course of [35S]methionine incorporation was performed at 37°C for periods up to 80 min. The selected experimental time for each mRNA was chosen as a time point in which linear incorporation of [35S]methionine was still occurring, essentially 80% of the linear range and is considered to be the optimum time (see ). [35S]methionine incorporation was monitored as hot, trichloroacetic acid (TCA) precipitable radioactivity and the precipitated radioactivity was quantitated using liquid scintillation spectroscopy.
For the start site selection experiments, reaction mixtures were incubated with the indicated amounts of mRNA and for an optimal time () and then stopped by placing the reaction mixture on ice. Five microliters were taken for the determination of [35S]methionine incorporation as hot TCA precipitable radioactivity and 10 μl of the reaction was mixed with 3 μl of 5X SDS sample buffer, heated to 95°C for 5 min and then radioactive proteins were resolved by SDS gel electrophoresis as described by Laemmli.Citation32 Following electrophoresis, the gels were dried and the radioactive bands visualized using a phosphImager. Densitometry of the individual protein bands was achieved using ImageQuant software. For all samples, the absolute amount of synthesis of each protein band was determined, but for ease in analysis, plots quantitating start site selection were plotted as percentage of the total (i.e., for each data point, the amount of initiation from start site 1 and 2 would total 100%).
When inhibitors were added to the reticulocyte lysates, they were pre-incubated with the entire reaction mixture minus the mRNA for 15 min. at 30°C. After this pre-incubation, mRNA was added at the optimal level followed by incubation for the amount of time optimized above. Inhibitors were added to the 25 μl reactions in the following amounts (where as concentrations, these are the final concentrations): m7GTP, 100 μM; Pdcd4, 0.6 μg; human p56, 180 nM; mouse p56, 122 nM; poly(I:C), 600 pg. Control reactions were treated in the same manner except no inhibitor was added. Except for m7GTP which barely inhibitied protein synthesis, the addition of the remaining inhibitors reduced [35S]methionine incorporation by 40 to 70% (data not shown).
All of the experiments described above were performed three or more times. The values shown in the figures represent the average of these experiments (with the standard deviation shown as well).
Synthesis of mRNAs – Capped mRNAs were made using T7 RNA polymerase, the “antireverse cap analog” (ARCA) version of m7GTP and plasmids containing the mRNA constructs as described by Ambion. The mRNAs were transcribed from plasmids provided by: Dr. Stan Tahara (University of Southern California) – rGB456 and T7CAT34;Citation31 Dr. Christopher Saris (Netherlands Cancer Center in Amsterdam) – Pim-2;Citation37,Citation48 Dr. Joseph Curran (University of Geneva) – pGEMP/C;Citation36 Dr. Richard Lloyd (Baylor College of Medicine) – eIF4G;Citation39 Dr. Anne-Catherine Prats (Centre Hospitalier Universitaire Rangueil) – cMYCCATP2.Citation38 The ratio of GTP to the ARCA m7GTP was 1 to 8. Following synthesis, the mRNAs were extracted with phenol and then precipitated with 70% ethanol. Precipitated mRNA was dissolved in nuclease free water and precipitated a second time with 70% ethanol. This precipitate was collected by centrifugation, briefly air-dried and then dissolved in nuclease free water at a concentration of 2 to 10 A260/ml (or 80 to 400 μg/ml). The final mRNA concentration was determined spectrally with a UV scan from 220 to 320 nm. mRNAs characterized by this methodology had the following spectral ratios: A260/A220 = 1, A260/A230 = 2, and A260/A280 = 2. It was assumed that 1 A260 of RNA was equal to 40 μg of RNA.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Drs. Thomas Dever (NIH) and Jon Lorsch (Johns Hopkins School of Medicine) for their review of the manuscript and many helpful comments.
References
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 1987; 196:947 - 50; http://dx.doi.org/10.1016/0022-2836(87)90418-9; PMID: 3681984
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol 1989; 9:5134 - 42; PMID: 2601712
- Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A 1990; 87:8301 - 5; http://dx.doi.org/10.1073/pnas.87.21.8301; PMID: 2236042
- Castilho-Valavicius B, Yoon H, Donahue TF. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics 1990; 124:483 - 95; PMID: 2179049
- Yoon HJ, Donahue TF. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol 1992; 12:248 - 60; PMID: 1729602
- Huang HK, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev 1997; 11:2396 - 413; http://dx.doi.org/10.1101/gad.11.18.2396; PMID: 9308967
- Fletcher CM, Pestova TV, Hellen CU, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J 1999; 18:2631 - 7; http://dx.doi.org/10.1093/emboj/18.9.2631; PMID: 10228174
- Battiste JL, Pestova TV, Hellen CU, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell 2000; 5:109 - 19; http://dx.doi.org/10.1016/S1097-2765(00)80407-4; PMID: 10678173
- Taylor JT, Devkota B, Huang AD, Topf M. Narayanan, E Sali, A Harvey SC Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Cell Structure 2009; 17:1591 - 604; http://dx.doi.org/10.1016/j.str.2009.09.015
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 2011; 331:730 - 6; http://dx.doi.org/10.1126/science.1198308; PMID: 21205638
- Zheng A, Yamamoto R, Sokabe M, Tanaka I, Yao M. Crystallization and preliminary X-ray crystallographic analysis of eIF5BΔN and the eIF5BΔN-eIF1AΔN complex. Acta Crystallogr Sect F Struct Biol Cryst Commun 2011; 67:730 - 3; http://dx.doi.org/10.1107/S1744309111015910; PMID: 21636924
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, et al. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep 2012; 1:689 - 702; http://dx.doi.org/10.1016/j.celrep.2012.04.007; PMID: 22813744
- Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 1998; 394:854 - 9; http://dx.doi.org/10.1038/29703; PMID: 9732867
- Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 2005; 20:251 - 62; http://dx.doi.org/10.1016/j.molcel.2005.09.008; PMID: 16246727
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 2006; 20:624 - 36; http://dx.doi.org/10.1101/gad.1397906; PMID: 16510876
- Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol 2009; 385:491 - 506; http://dx.doi.org/10.1016/j.jmb.2008.10.029; PMID: 18976658
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 2004; 18:3078 - 93; http://dx.doi.org/10.1101/gad.1255704; PMID: 15601822
- Cheung Y-N, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, et al. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo.. Genes Dev 2007; 21:1217 - 30; http://dx.doi.org/10.1101/gad.1528307; PMID: 17504939
- Alone PV, Cao C, Dever TE. Translation initiation factor 2γ mutant alters start codon selection independent of Met-tRNA binding. Mol Cell Biol 2008; 28:6877 - 88; http://dx.doi.org/10.1128/MCB.01147-08; PMID: 18794367
- Takacs JE, Neary TB, Ingolia NT, Saini AK, Martin-Marcos P, Pelletier J, et al. Identification of compounds that decrease the fidelity of start codon recognition by the eukaryotic translational machinery. RNA 2011; 17:439 - 52; http://dx.doi.org/10.1261/rna.2475211; PMID: 21220547
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 2012; 40:2898 - 906; http://dx.doi.org/10.1093/nar/gkr1192; PMID: 22156057
- ElAntak L, Wagner S, Herrmannova A, Karaskova M, Rutkai E. Lukavsky Valasek L. The indispensable N-terminal half of eIF3j/HCR1 cooperates with is structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J Mol Biol 2010; 396:1097 - 116; http://dx.doi.org/10.1016/j.jmb.2009.12.047; PMID: 20060839
- Martin-Marcos P, Cheung YN, Hinnebusch AG. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol 2011; 31:4814 - 31; http://dx.doi.org/10.1128/MCB.05819-11; PMID: 21930786
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev 2012; 24:97 - 110; http://dx.doi.org/10.1101/gad.1871910
- Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 2011; 75:434 - 67; http://dx.doi.org/10.1128/MMBR.00008-11; PMID: 21885680
- Asano K, Sachs MS. Translation factor control of ribosome conformation during start codon selection. Genes Dev 2007; 21:1280 - 7; http://dx.doi.org/10.1101/gad.1562707; PMID: 17545463
- Hinnebusch AG. Dever TE Asano K. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In Translational Control in Biology and Medicine (ed. M. B. Mathews, N. Sonenberg and J. H. B. Hershey), Cold Spring Harbor, NY:Cold Spring Harbor Laboratory Press, 2007:225-268.
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 2012; 19:568 - 76; http://dx.doi.org/10.1038/nsmb.2303; PMID: 22664984
- Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev 2000; 14:2534 - 46; http://dx.doi.org/10.1101/gad.831800; PMID: 11018020
- Merrick WC. Initiation of protein biosynthesis in eukaryotes. Bioc. Mol. Biol. Ed. 2003; 31:378-385.
- Tahara SM, Dietlin TA, Dever TE, Merrick WC, Worrilow LM. Effect of eukaryotic initiation factor 4F on AUG selection in a bicistronic mRNA. J Biol Chem 1991; 266:3594 - 601; PMID: 1995620
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680 - 5; http://dx.doi.org/10.1038/227680a0; PMID: 5432063
- Hui DJ, Terenzi F, Merrick WC, Sen GC. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J Biol Chem 2005; 280:3433 - 40; http://dx.doi.org/10.1074/jbc.M406700200; PMID: 15561726
- Hui DJ, Bhasker CR, Merrick WC, Sen GC. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J Biol Chem 2003; 278:39477 - 82; http://dx.doi.org/10.1074/jbc.M305038200; PMID: 12885778
- Yang H-S, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 2003; 23:26 - 37; http://dx.doi.org/10.1128/MCB.23.1.26-37.2003; PMID: 12482958
- Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol Cell Biol 1998; 18:5021 - 31; PMID: 9710586
- Saris CJ, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J 1991; 10:655 - 64; PMID: 1825810
- Nanbru C, Lafon I, Audigier S, Gensac MC, Vagner S, Huez G, et al. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem 1997; 272:32061 - 6; http://dx.doi.org/10.1074/jbc.272.51.32061; PMID: 9405401
- Byrd MP, Zamora M, Lloyd RE. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J Biol Chem 2005; 280:18610 - 22; http://dx.doi.org/10.1074/jbc.M414014200; PMID: 15755734
- Adams SL, Safer B, Anderson WF, Merrick WC. Eukaryotic initiation complex formation. Evidence for two distinct pathways. J Biol Chem 1975; 250:9083 - 9; PMID: 1194278
- Merrick WC. Evidence that a single GTP is used in the formation of 80 S initiation complexes. J Biol Chem 1979; 254:3708 - 11; PMID: 438155
- Peterson DT, Safer B, Merrick WC. Role of eukaryotic initiation factor 5 in the formation of 80 S initiation complexes. J Biol Chem 1979; 254:7730 - 5; PMID: 468782
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci U S A 2010; 107:18056 - 60; http://dx.doi.org/10.1073/pnas.1009269107; PMID: 20921384
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 2011; 39:7598 - 609; http://dx.doi.org/10.1093/nar/gkr484; PMID: 21705780
- Das S, Ghosh R, Maitra U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J Biol Chem 2001; 276:6720 - 6; http://dx.doi.org/10.1074/jbc.M008863200; PMID: 11092890
- Komar AA, Mazumder B. Merrick WC. A new framework for understanding IRES-mediated translation. Gene 2012; 10:75 - 86; http://dx.doi.org/10.1016/j.gene.2012.04.039
- Raught B. Gingas A-C. Signaling to translation initiation. In Translational Control in Biology and Medicine (ed. M. B. Mathews, N. Sonenberg and J. H. B. Hershey), Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press, 2007:369-400.
- van der Lugt NMT, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, et al. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J 1995; 14:2536 - 44; PMID: 7781606
- Benne R, Brown-Luedi ML, Hershey JW. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J Biol Chem 1978; 253:3070 - 7; PMID: 641055
- Thomas A, Goumans H, Amesz H, Benne R, Voorma HO. A comparison of the initiation factors of eukaryotic protein synthesis from ribosomes and from the postribosomal supernatant. Eur J Biochem 1979; 98:329 - 37; http://dx.doi.org/10.1111/j.1432-1033.1979.tb13192.x; PMID: 488105
- Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol 1977; 116:755 - 67; http://dx.doi.org/10.1016/0022-2836(77)90269-8; PMID: 592399
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 2000; 403:332 - 5; http://dx.doi.org/10.1038/35002118; PMID: 10659855
- Staehelin T, Erni B, Schreier MH. Purification and characterization of seven initiation factors for mammalian protein synthesis. Methods Enzymol 1979; 60:136 - 65; http://dx.doi.org/10.1016/S0076-6879(79)60013-7; PMID: 459895
- Safer B, Anderson WF, Merrick WC. Purification and physical properties of homogeneous initiation factor MP from rabbit reticulocytes. J Biol Chem 1975; 250:9067 - 75; PMID: 1194277
- Merrick WC, Kemper WM, Anderson WF. Purification and characterization of homogeneous initiation factor M2A from rabbit reticulocytes. J Biol Chem 1975; 250:5556 - 62; PMID: 1095581
- Merrick WC, Anderson WF. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem 1975; 250:1197 - 206; PMID: 1112800
- Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte initiation factor M2Bα and M2Bβ. J Biol Chem 1976; 251:5551 - 7; PMID: 965377
- Safer B, Adams SL, Kemper WM, Berry KW, Lloyd M, Merrick WC. Purification and characterization of two initiation factors required for maximal activity of a highly fractionated globin mRNA translation system. Proc Natl Acad Sci U S A 1976; 73:2584 - 8; http://dx.doi.org/10.1073/pnas.73.8.2584; PMID: 1066667
- Grifo JA, Tahara SM, Leis JP, Morgan MA, Shatkin AJ, Merrick WC. Characterization of eukaryotic initiation factor 4A, a protein involved in ATP-dependent binding of globin mRNA. J Biol Chem 1982; 257:5246 - 52; PMID: 7068683
- Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem 1983; 258:5804 - 10; PMID: 6853548
- Merrick WC. Assays for eukaryotic protein synthesis. Methods Enzymol 1979; 60:108 - 23; http://dx.doi.org/10.1016/S0076-6879(79)60011-3; PMID: 459892
- Merrick WC, Barth-Baus D. Use of reticulocyte lysates for mechanistic studies of eukaryotic translation initiation. Methods Enzymol 2007; 429:1 - 21; http://dx.doi.org/10.1016/S0076-6879(07)29001-9; PMID: 17913616