Abstract
In the budding yeast Saccharomyces cerevisiae, the cyclin-dependent kinases (CDKs) Kin28, Bur1 and Ctk1 regulate basal transcription by phosphorylating the carboxyl-terminal domain (CTD) of RNA polymerase II. However, very little is known about the involvement of the cell cycle CDK Cdc28 in the transcription process. We have recently shown that, upon cell cycle entry, Cdc28 kinase activity boosts transcription of a subset of genes by directly stimulating the basal transcription machinery. Here, we discuss the biological significance of this finding and give our view of the kinase-dependent role of Cdc28 in regulation of RNA polymerase II.
Introduction
Transcription of eukaryotic genes by RNA polymerase II (RNAP II) is a highly regulated stepwise mechanism involving hundreds of proteins. The initial step is the assembly of general transcription factors (TFII-A, -B, -D, -E and -H) and RNAP II to form the pre-initiation complex (PIC). The CTD of Rpb1, RNAP II largest subunit, then undergoes sequential phosphorylations by the cyclin-dependent kinases (CDKs) Kin28, Bur1 and Ctk1 (respectively CDK7, CDK9 and CDK12 in humans).Citation1,Citation2 These phosphorylation events allow RNAP II to start mRNA synthesis, but also to couple transcription to mRNA processing and chromatin modification during transcription elongation and termination.Citation1
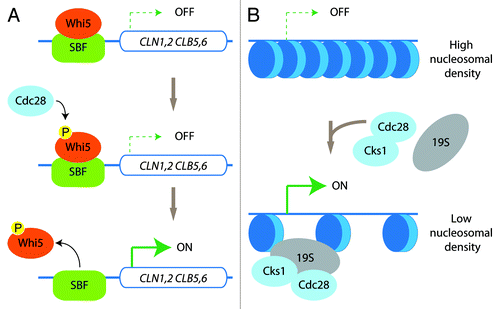
Transcription initiation is also regulated by sequence-specific DNA-binding transcription factors that are differentially recruited to specific subsets of genes to allow their expression to occur at the right place and at the right moment.Citation3 Such temporal regulation of gene expression has been well characterized during cell cycle progression.Citation4 In S. cerevisiae, the CDK Cdc28 (also known as Cdk1) is necessary and sufficient for cell cycle regulation by phosphorylating a large number of substrates to coordinate cell cycle events.Citation5 Specifically, Cdc28 phosphorylates Whi5 in late G1, leading to dissociation of Whi5 from the transcription factor complex SBF (Swi4/6-dependent cell-cycle box binding factor). Dissociation of Whi5 activates SBF, which then induces expression of a group of genes (including cyclin genes CLN1,2 and CLB5,6) to induce cell cycle entryCitation6 ().
Figure 1. Kinase-dependent and -independent roles of Cdc28. (A) In late G1, Cdc28 phosphorylates the SBF-bound repressor Whi5. This triggers the release of Whi5 from the SBF, thus stimulating the transcription of the genes belonging to the G1 transcriptional program. (B) Cdc28/Cks1 and the 19S-regulatory particle of the proteasome are recruited to genes such as GAL1 for efficient transcriptional activation by promoting a decrease in nucleosomal density.
We have recently shown that Cdc28 also boosts the expression of a specific subset of housekeeping genes (that do not contain any SBF consensus sequence) by directly stimulating the recruitment of RNAP II and Kin28, and by promoting CTD phosphorylation and mRNA capping.Citation7 These genes encode proteins involved in energy supply, translation, cell wall integrity and chromatin architecture. We propose that upon cell cycle entry Cdc28 directly stimulates the basal transcription machinery at these genes in order to maintain sufficiently high expression levels of these proteins as the bud grows and total cell volume increases. These findings highlight a new role for Cdc28 as a cell cycle-regulated transcriptional CDK and raise important mechanistic questions about the sequence of events and the identity of the factors involved in Cdc28 recruitment at these particular loci.
The CTD kinases involved in the CTD phosphorylation cycle
The CTD of Rpb1 is characterized by multiple repeats of the heptapeptide Y1S2P3T4S5P6S7 (52 repeats in humans and 26 in yeast). In yeast, Y1, S2, S5 and S7 have been found to be phosphorylated during transcription, while mammals in addition also phosphorylate T4.Citation1,Citation8,Citation9 Early in the transcription cycle, Kin28 phosphorylates the CTD on S5, which serves as a mark for recruitment of the mRNA capping machinery.Citation10 As RNAP II elongates, the levels of phosphorylated S5 progressively decrease due to the action of the phospho-S5-specific phosphatases Rtr1 and Ssu72, while the phosphorylation of S2 increases toward the 3′ end of the ORF due to activity of Bur1 and Ctk1. Phosphorylated S2 serves as a docking site for a multitude of protein complexes involved in histone modification, chromatin remodeling, mRNA polyadenylation and transcription termination.Citation1 S2 and S5 can also be phosphorylated by Srb10 (CDK8 in humans), the catalytic subunit of the Mediator’s kinase module.Citation11 In vitro studies showed that, on the one hand, Srb10 inhibits transcription by preventing PIC formation, but on the other hand, it stimulates transcription by promoting the formation of the scaffold complex.Citation12 Phosphorylation of S7 is performed by Kin28 and Bur1, and contributes to expression of ncRNA and mRNA splicing.Citation13-Citation15 Finally, in mammals, Y1 phosphorylation, possibly by the tyrosine kinase c-Abl, promotes transcription terminationCitation9,Citation16. Y1 phosphorylation was recently also demonstrated in S. cerevisiaeCitation9; however, the kinase remains unknown and c-Abl does not exist in yeast. The CTD kinase activities of Ctk1, Kin28, Srb10 and Bur1 were characterized between 1991 and 1995,Citation17 but the very first kinase shown to phosphorylate the CTD was murine CDC2 (the ortholog of Cdc28).Citation18 Quite surprisingly, the function of CDC2/Cdc28 in regulation of basal transcription has only been investigated sporadically, and the physiological consequences have largely remained obscure.
Cdc28 kinase activity promotes transcription in a cell cycle-dependent manner
An early study that probed the involvement of Cdc28 in basal transcription found that the recruitment of the Mediator complex at SBF-regulated genes (such as HO or CLN1/2) requires a certain basal level of Cdc28 activity, and that high Cdc28 activity is necessary for subsequent recruitment of RNAP II, TFIIH and TFIIB.Citation19 However, the requirement for Cdc28 in transcription of these genes may in fact be to remove Whi5Citation6 rather than directly activating the basal transcription machinery.
Interestingly, more recent studies identified a kinase-independent role for Cdc28 in basal transcription. It was proposed that Cdc28 and its interaction partner Cks1 were recruited to the promoter of CDC20 (a regulator of the anaphase promoting complex), to regulate the periodic association of the proteasome.Citation20,Citation21 Cdc28/Cks1 also recruit the proteasome to GAL1, which mediates efficient nucleosome evictionCitation22,Citation23 ().
These studies focused on a limited number of model genes, while in our recent report we aimed at determining all the genomic binding sites of Cdc28. Using ChIP followed by high throughput sequencing (ChIP-seq) we found that Cdc28 could be detected at approximately 2,000 ORFs, with the 10th percentile showing at least a 3-fold recruitment over background.Citation7 Interestingly, these 200 ORFs belong to most highly transcribed genes in yeast, e.g., PMA1. To dissect the involvement of Cdc28 in basal transcription, we then used a mutant strain harboring the cdc28-as1 allele, which encodes a form of Cdc28 that is highly sensitive to the inhibitor 1-NM-PP1,Citation24 and found that PMA1 transcription depends on Cdc28 kinase activity. Furthermore, we found that PMA1 mRNA levels were low in G1 phase (when Cdc28 activity is low) and peaked upon entry into the cell cycle, when Cdc28 becomes active.Citation7 Together, these data strongly suggest that Cdc28 stimulates transcription of PMA1 during cell cycle entry.
The function of the kinase activity of Cdc28 in transcription is not mutually exclusive with the previously described, non-catalytic role of Cdc28 in this process. The relative contribution of both these functions of Cdc28 in regulation of transcription remains to be determined.
Is Cdc28 a cell cycle-regulated CTD kinase?
How does Cdc28 boost transcription of its target genes during cell cycle entry? A previous report indicated that human CDC2 can phosphorylate both S2 and S5 in vitro,Citation25 and we first confirmed that Cdc28 can also directly phosphorylate the CTD. However, our experiments suggest that Cdc28 preferentially targets S5 in vitro and that it has little or no activity toward either S2 or S7.Citation7 These findings were supported by in vivo results obtained with ChIP assays and western blots, which also showed that Cdc28 cooperates with Kin28 to achieve full S5 phosphorylation and to promote mRNA capping.Citation7 These data suggest that Cdc28 is a CTD kinase sharing a partially redundant role with Kin28 in CTD-S5 phosphorylation.
Mutual regulation of Cdc28 and Kin28
How is Cdc28 recruited to RNAP II? Recently, Kin28-mediated phosphorylation of CTD-S5 was shown to serve as a priming site for Bur1 recruitment.Citation26 Interestingly, it was shown that Cdc28 is also regulated by a priming mechanism; in particular, Cks1 serves as a phospho-acceptor binding site that recruits the Cdc28 holoenzyme to at least some of its substrates.Citation27
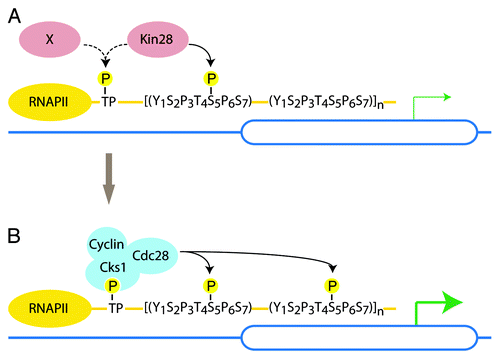
These two studies suggest that recruitment of Cdc28 to the CTD may similarly depend upon CTD priming. Indeed, our ChIP assays showed that full Kin28 kinase activity is necessary for the recruitment of Cks1/Cdc28 to PMA1.Citation7 Consistently, we found that human CDK7 can prime the CTD for further phosphorylation by Cks1/Cdc28 in vitro (our unpublished data and Dr. M. Loog, personal communication). Thus, we propose that recruitment of Cdc28 to RNAP II may depend on priming of the CTD by Kin28 ().
Figure 2. Model for Cdc28-mediated phosphorylation of the CTD. (A) In late G1 Kin28 (or potentially a still uncharacterized kinase) primes the CTD. (B) Phosphorylated TP sites (threonine followed by a proline) and/or multiple phospho-S5 serve as a docking site for Cks1/Cdc28. As Cdc28 becomes active in late G1, Cdc28 further phosphorylates S5 to ensure high rates of transcription and mRNA capping.
Conclusions
Our recent study suggests that direct regulation of CTD-S5 phosphorylation may serve as a switch to regulate the basal transcription machinery during the cell cycle. However, the sequence of events by which Cdc28 stimulates transcription and mRNA capping remains unclear. Although our study revealed a functional interaction between Kin28 and Cdc28, Cdc28 may have additional roles in the transcription process. Indeed, we found that CDC28 genetically interacts with BUR2 and CTK1 and that Cdc28 is recruited to the entire ORF of its target genes, suggesting it also has a role in transcription elongation.Citation7
With the recent discovery of new CTD kinases like Cdc28, CDK12 and PLK, and the discovery of CTD-Y1 and CTD-T4 phosphorylation, the CTD phosphorylation code appears much more complex than previously anticipated.Citation2,Citation8,Citation9 These CTD modifications allow the cell to integrate a large variety of signals, thereby fine-tuning transcription during a host of biological processes like development and cell division.
| Abbreviations: | ||
| RNAP II | = | RNA polymerase II |
| PIC | = | pre-initiation complex |
| CTD | = | Carboxyl-terminal domain |
| CDK | = | cyclin-dependent kinase |
| SBF | = | Swi4/6-dependent cell-cycle box binding factor |
| ORF | = | open reading frame |
Acknowledgments
We would like to thank Mart Loog for critically reading the manuscript. JME is supported by the Norwegian Research Council (YFF award #180499) and by the Norwegian Health Authority South-East (grants #2010036 and 2012012).
References
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell 2009; 36:541 - 6; http://dx.doi.org/10.1016/j.molcel.2009.10.019; PMID: 19941815
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 2010; 24:2303 - 16; http://dx.doi.org/10.1101/gad.1968210; PMID: 20952539
- Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 2000; 14:2551 - 69; http://dx.doi.org/10.1101/gad.831000; PMID: 11040209
- Simmons Kovacs LA, Mayhew MB, Orlando DA, Jin Y, Li Q, Huang C, et al. Cyclin-dependent kinases are regulators and effectors of oscillations driven by a transcription factor network. Mol Cell 2012; 45:669 - 79; http://dx.doi.org/10.1016/j.molcel.2011.12.033; PMID: 22306294
- Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div 2010; 5:11; http://dx.doi.org/10.1186/1747-1028-5-11; PMID: 20465793
- Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 2005; 24:2746 - 55; http://dx.doi.org/10.1038/sj.onc.1208606; PMID: 15838511
- Chymkowitch P, Eldholm V, Lorenz S, Zimmermann C, Lindvall JM, Bjørås M, et al. Cdc28 kinase activity regulates the basal transcription machinery at a subset of genes. Proc Natl Acad Sci U S A 2012; 109:10450 - 5; http://dx.doi.org/10.1073/pnas.1200067109; PMID: 22689984
- Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J 2012; 31:2784 - 97; http://dx.doi.org/10.1038/emboj.2012.123; PMID: 22549466
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012; 336:1723 - 5; http://dx.doi.org/10.1126/science.1219651; PMID: 22745433
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 2000; 20:104 - 12; http://dx.doi.org/10.1128/MCB.20.1.104-112.2000; PMID: 10594013
- García A, Rosonina E, Manley JL, Calvo O. Sub1 globally regulates RNA polymerase II C-terminal domain phosphorylation. Mol Cell Biol 2010; 30:5180 - 93; http://dx.doi.org/10.1128/MCB.00819-10; PMID: 20823273
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev 2005; 19:1401 - 15; http://dx.doi.org/10.1101/gad.1318105; PMID: 15964991
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 2007; 318:1780 - 2; http://dx.doi.org/10.1126/science.1145977; PMID: 18079404
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 2009; 34:387 - 93; http://dx.doi.org/10.1016/j.molcel.2009.04.016; PMID: 19450536
- Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007; 318:1777 - 9; http://dx.doi.org/10.1126/science.1145989; PMID: 18079403
- Baskaran R, Escobar SR, Wang JY. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the mammalian RNA polymerase II: possible role in transcription elongation. Cell Growth Differ 1999; 10:387 - 96; PMID: 10392900
- Bartkowiak B, Greenleaf AL. Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb?. Transcription 2011; 2:115 - 9; http://dx.doi.org/10.4161/trns.2.3.15004; PMID: 21826281
- Cisek LJ, Corden JL. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature 1989; 339:679 - 84; http://dx.doi.org/10.1038/339679a0; PMID: 2662013
- Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell 2001; 7:1213 - 20; http://dx.doi.org/10.1016/S1097-2765(01)00266-0; PMID: 11430824
- Arvai AS, Bourne Y, Hickey MJ, Tainer JA. Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain. J Mol Biol 1995; 249:835 - 42; http://dx.doi.org/10.1006/jmbi.1995.0341; PMID: 7791211
- Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 2003; 423:1009 - 13; http://dx.doi.org/10.1038/nature01720; PMID: 12827207
- Chaves S, Baskerville C, Yu V, Reed SI. Cks1, Cdk1, and the 19S proteasome collaborate to regulate gene induction-dependent nucleosome eviction in yeast. Mol Cell Biol 2010; 30:5284 - 94; http://dx.doi.org/10.1128/MCB.00952-10; PMID: 20855529
- Yu VP, Baskerville C, Grünenfelder B, Reed SI. A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol Cell 2005; 17:145 - 51; http://dx.doi.org/10.1016/j.molcel.2004.11.020; PMID: 15629725
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 2000; 407:395 - 401; http://dx.doi.org/10.1038/35030148; PMID: 11014197
- Zhang J, Corden JL. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem 1991; 266:2290 - 6; PMID: 1899239
- Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell 2009; 33:752 - 62; http://dx.doi.org/10.1016/j.molcel.2009.02.018; PMID: 19328068
- Kõivomägi M, Valk E, Venta R, Iofik A, Lepiku M, Balog ER, et al. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 2011; 480:128 - 31; http://dx.doi.org/10.1038/nature10560; PMID: 21993622