Abstract
Exonuclease (exo) III was used as a probe of the Escherichia coli RNA polymerase (RNAP) ternary elongation complex (TEC) downstream border. In the absence of NTPs, RNAP appears to stall primarily in a post-translocated state and to return slowly to a pre-translocated state. Exo III mapping, therefore, appears inconsistent with an unrestrained thermal ratchet model for translocation, in which RNAP freely and rapidly oscillates between pre- and post-translocated positions. The forward translocation state is made more stable by lowering the pH and/or by elevating the salt concentration, indicating a probable role of protonated histidine(s) in regulating accurate NTP loading and translocation. Because the post-translocated TEC can be strongly stabilized by NTP addition, NTP analogs were ranked for their ability to preserve the post-translocation state, giving insight into RNAP fidelity. Effects of NTPs (and analogs) and analysis of chemically modified RNA 3′ ends demonstrate that patterns of exo III mapping arise from intrinsic and subtle alterations at the RNAP active site, far from the site of exo III action.
Introduction
The nucleic acid scaffolds for RNA polymerase (RNAP) pre- and post-translocated ternary elongation complexes (TECs) are depicted in . RNAP appears to rest in a primarily post-translocated state and to return to the pre-translocated state slowly and reversibly.Citation1,Citation2 This conclusion has been advanced based on X-ray crystal structures of RNAP TECs, which are mostly post-translocated.Citation3-Citation8 Fluorescence changes associated with translocation also indicate this conclusion,Citation1 as do exonuclease (exo) III mapping studies of TEC boundaries.Citation2 Here, we support this model by tuning the exo III mapping experiment and stabilizing the post-translocation state, using varied RNA length, lowered pH and salt in the physiological range. Multi-subunit RNAPs do not appear to rapidly oscillate between pre- and post-translocation positions. The transition from the pre- to the post-translocated state must be rapid, to account for rapid elongation rates; but as we show here, the return to the pre-translocated state is much slower and tunable by strengthening or weakening RNAP-nucleic acid interactions.
Figure 1. A 9 nt RNA + NTP length gives very strong NTP stabilization of the post-translocation state of the RNAP TEC. (A) Nucleotide scaffolds for pre- and post-translocated TECs (PRE and PST). Template DNA strand (TDS) is blue; non-template DNA strand (NDS) is green; RNA is red. The NTP substrate (red) is in stick representation. Mg2+ is magenta. The closed trigger loop (TL) is yellow. β’ H936 is cyan. The image was derived from PDB 205JCitation5 and drawn using Visual Molecular Dynamics.Citation38 (B) Schematic of experiments for downstream border exo III mapping at TEC-G8 and TEC-A9. * indicates a 32P radiolabel; # indicates a sulfur for oxygen substitution in the TDS to block exo III (orange) digestion. Arrows indicate the upstream to downstream direction of transcription. The positions of the i and i+1 sites are indicated for pre- and post-translocated TECs. At 40 mM KCl, exo III digestion is blocked primarily at the i+18 position. At higher KCl and/or lower pH, digestion can be slowed at i+19 and i+18 (see below). As in panel A, the TDS is blue, the NDS is green and the RNA is red. The TEC bubble is indicated in outlined letters and pink shading. (C) Effects of NTPs (100 μM ATP or CTP) on chain-terminated 3′dG8 and 3′dA9 TECs. Exo III reaction times are in seconds (s). (D) Translocation of G7, G8 and A9 TECs (no chain termination). KCl is 40 mM; pH is 7.9. Backtracked (BTR), pre- (PRE) and post-translocated (PST) TECs are indicated.
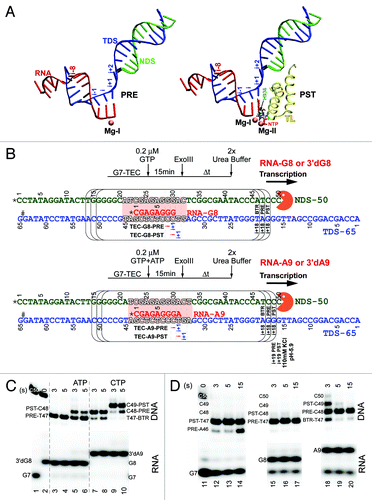
The RNAP elongation mechanism is expected to involve deprotonation of the 3′-OH of the RNA (i site; post-translocated TEC) to enhance nucleophilic attack on the α-phosphate of the substrate NTP (i+1 site).Citation9-Citation11 A critical histidine (β’ H936; Escherichia coli RNAP numbering) on the mobile “trigger loop” is expected to become protonated to interact with the NTP substrate β-phosphate (i+1 site) (; right panel). H936 is thought to transfer this proton to the β-phosphate at the time of chemistry to improve pyrophosphate (PPi or PPiH) as a leaving group. Such a mechanism might be expected to be general acid-base catalyzed because H936 protonation is favored by a lower pH environment and because 3′-OH deprotonation is enhanced in a higher pH environment. It should be noted that proton transfer from H936 cannot be obligatory for phosphodiester bond formation because substitutions of H936 and homologous positions in other RNAPs can be active, demonstrating that alternate mechanisms are possible.Citation12-Citation14 Furthermore, RNAP elongation appears to be general base catalyzedCitation13 indicating that the polymerization reaction is dominated by deprotonation of the 3′-OH and appears less reliant on proton transfer from the trigger loop H936. By dividing the RNAP reaction into translocation and NTP-loading phases, as we do here, we reveal acid and/or salt stimulated steps in the RNAP reaction that are independent of 3′-OH deprotonation. Indeed, 3′-OH deprotonation cannot occur in many of these exo III mapping experiments because we often use a 3′deoxy chain terminator to block the polymerization step, allowing the incoming NTP substrate to be added without incorporation. Significantly, both NTP-dependent and NTP-independent stabilization of forward RNAP translocation appear to be enhanced by lower pH and/or increased salt (into the physiological range).
Exo III attacks DNA in a 3′→5′ direction. In this paper, we digest non-template DNA to the downstream RNAP TEC boundary (). Attack by exo III, therefore, provides information about the transition from the post-translocated state to the pre-translocated state, and insight is also obtained into RNAP backtracking. Once the DNA is shortened by exo III, it remains shortened even when RNAP translocates forward. So, in this reaction format, translocation rates are only estimated in the downstream→upstream direction. Because of this limitation of the exo III assay, reversibility from pre-→post-translocation states, although this may occur rapidly, cannot be observed. Post-→pre-translocation rates estimated in the exo III reaction are very slow compared with transcriptional elongation rates, indicating that, when depleted of NTPs, RNAP stalls predominantly in a post-translocated register.Citation2
Results and Discussion
Because the exo III mapping experiment gives an indirect readout and can be confusing, it is illustrated in detail in and . shows structural models for TEC nucleic acid scaffoldsCitation4,Citation5 that represent pre- and post-translocation states. The metric used for numbering TEC translocation registers is shown. The RNA/DNA hybrid (pre-) or the RNA+NTP/DNA hybrid (post-) length is 10 nt (i-8 to i+1). The TEC single-stranded DNA bubble is about 12–13 nts.Citation15,Citation16 shows a schematic representation of the exo III mapping experiment and indicates how exo III resistant DNA band patterns generated at the downstream TEC boundary are interpreted.Citation2 The active site of RNAP comprises the i and i+1 sites. In the pre-translocation state, for a RNA of length n, the n-1 RNA base is in the i site, and the n RNA base is in the i+1 site, so forward translocation must occur for NTP loading to the active site at i+1. In the post-translocation state, the n RNA base is in the i site, and the NTP substrate can occupy the i+1 site for catalysis (). For our experiments, the non-template C50 DNA strand (NDS-50), which is attacked and shortened by exo III, and the RNA are 5′-end labeled with 32P for detection on gels (). The G65 DNA template strand includes a sulfur for oxygen substitution at the 3′-end to block exo III digestion, so exo III only attacks the non-template DNA strand.Citation17 Exo III cleaves dCMP more rapidly than other bases,Citation17 so a NDS-50 strand with 48-CCC-50 was designed. Standard interpretations of exo III patterns are reported, with digestion primarily to the i+18 downstream positionCitation2,Citation17-Citation19 (at 40 mM KCl), but we emphasize that the exo III assay is an indirect readout of translocation (also, see discussion below). A strong argument for band assignments is that the exo III digestion patterns that we identify as resulting primarily from a post-translocated TEC are strongly stabilized by addition of accurately templated NTPs (). The exo III patterns identified as resulting from a pre-translocated TEC, as expected, are shifted by one nt upstream, resulting in the NDS being shortened by one additional nt.
A potent effect of the incoming NTP on RNAP translocation has rarely been observed in exo III mapping experiments.Citation2,Citation17,Citation18,Citation20 By adjusting the length of the RNA, however, the response to cognate NTPs was enhanced, making exo III a more useful probe for the RNAP translocation state and for NTP loading (Figs. One and S1). Because NTP stabilization of forward translocation is robust (), a spectrum of NTP analogs could be tested for their ability to stabilize forward translocation (see and below). We previously showed that, for E. coli RNAP using exo III, a 10 nt RNA tends to be detected in a pre-translocated state, and, by comparison, a 11 nt RNA tends to backtrack more readily by one nt2. These results are consistent with X-ray crystal structures because, in Thermus thermophilus RNAP TECs, the RNA+NTP/DNA hybrid length is 10 ntCitation4,Citation5 (). We reasoned, therefore, that a 10 nt RNA precisely fits the 10 nt channel hybrid length observed in structures, and therefore the TEC with a 10 nt RNA was observed primarily in the pre-translocated state. The TEC with a 11 nt RNA, by contrast, tended to backtrack by one nt to fill the 10 nt channel length, probably because a single nt was insufficient to stably occupy the RNA exit channel, allowing more rapid upstream sliding. We therefore considered the idea that exo III mapping experiments might be rendered more strongly NTP dependent if done on scaffolds with shorter RNA lengths that tend to resist backtracking and that may be more sensitive to cognate NTP addition.
Figure 3. Exo III mapping to obtain a ranking of GTP analogs (400 μM) to stabilize forward RNAP TEC translocation at 40 (upper panel) and 150 mM KCl (lower panel). PPi was at 1 mM. pH is at 7.9.
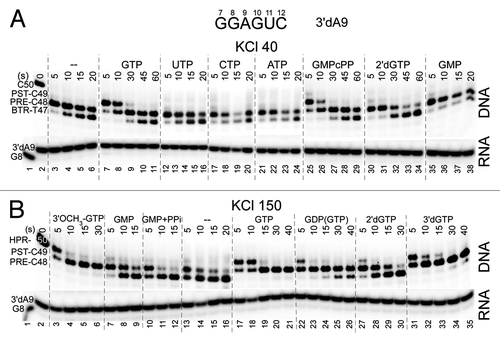
Figure 4. Exo III mapping to obtain a ranking of CTP analogs (400 μM) to stabilize forward RNAP TEC translocation at 40 mM KCl and pH 7.9.
From optimization experiments (Figs. One and S1), a TEC with a 9 nt RNA with a 3′-deoxy AMP chain terminator (3′dA9) was found to provide a robust response to NTP addition. In , chain terminated 3′dG8 and 3′dA9 TECs were compared for their responses to templated ATP and CTP substrates. A robust response to NTPs was observed for both templates, but the response of 3′dA9 to CTP addition appeared to be stronger (; compare lanes 3 and 4 (no ATP) with 5 and 6 (+ 100 μM ATP) and lanes 7 and 8 (no CTP) with 9 and 10 (+ 100 μM CTP)). Figure S1 shows similar experiments using RNAs of lengths 10–13 nt, for comparison. These experiments show that RNA lengths from 8–11 nt appear most useful for exo III mapping of RNAP TECs using what were our standard reaction conditions (transcription buffer with pH 7.9 and 40 mM KCl).Citation2 RNAs of lengths 12–13 nt generally induce much more backward slipping than shorter RNAs of lengths 8–11 nt and, probably for this reason, show much weaker responses to NTPs, indicating that RNA lengths of 8–11 nt may be preferable for exo III mapping experiments to observe translocation states and the fidelity of NTP loading. As we show below, however, adjusting the KCl and pH of buffers is a highly likely strategy to improve exo III mapping of TECs with longer RNAs, because these conditions strongly restrict upstream RNAP sliding. From our point of view, the 3′dA9 TEC is very useful for exo III mapping but, perhaps, ought not to be viewed as a particularly exceptional case. A 9 nt RNA length simply allows RNAP sliding to be slowed sufficiently to use exo III as a potent probe of translocation states in bench top experiments.
A9 RNA has the sequence 5′-CGAGAGGGA-3′. The tendency toward backtracking, therefore, can be attributed to the 3′-A, because a RNA with a 3′-G or 3′-C, for instance, would be expected to hold the forward translocation register more tenaciously.Citation2,Citation18,Citation21 Using this scaffold, CTP is the next templated NTP from A9 (), and 100 μM CTP strongly stabilizes forward translocation of the 3′-dA9 TEC (). The RNA controls indicate that termination at 3′dG8 and 3′dA9 was efficient, allowing barely detectable extension in the presence of ATP or CTP.
In , we show that, with a natural 3′- OH, the post-translocated A9 TEC can be observed at early times of incubation. The G8 TEC is mostly observed at the pre-translocated register within this time window (3–15 sec), and the G7 TEC appears to remain stubbornly post-translocated. Comparing the data in and , for the chain terminated 3′dA9 and natural A9 TECs [lanes 7 and 8 () and 18 and 19 ()], it appears that a RNA 3′-OH end may stabilize the post-translocated state slightly relative to a 3′-H2 chain-terminated end (also, see below). We conclude that the 3′dA9 TEC is highly suitable to observe strong NTP-dependent effects on RNAP TEC translocation, in order to preserve post-translocated TEC stability and to analyze transcriptional fidelity. Note that a 3′dA9 TEC + CTP just fills the 10 nt RNA+NTP/DNA hybrid channel ().
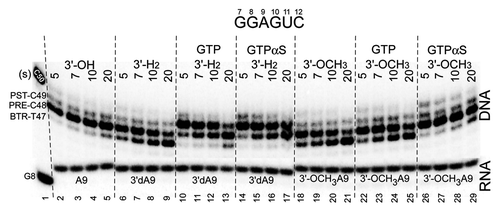
Figure 5. A natural 3′-OH RNA more stably maintains the post-translocation register of RNAP than 3′-H2 and 3′-OCH3 3′ ends, demonstrating the specificity of the exo III mapping assay for the RNAP TEC. GTPαS appears to stabilize the forward translocation state of the TEC slightly more strongly than GTP. GTP and GTPαS were added at 400 μM. The assay was at 40 mM KCl and pH 7.9.
pH and salt effects on translocation
Histidine protonation is increased by lowering pH and raising salt.Citation22 Elevating salt concentration is expected to support histidine protonation by stabilizing positive charge on the imidazole ring. Because a protonated H936 on the closed trigger loop might be expected to stabilize NTP binding by interaction with the β-phosphateCitation9-Citation11,Citation23 (; right panel), we considered the possibility that NTP-stabilized translocation might be stimulated by increasing salt and/or lowering pH. Here we show that the situation appears more complicated than a role for a protonated/deprotonated H936 in trigger loop dependent NTP loading and RNAP chemistry, because, somewhat surprisingly, both NTP-dependent and NTP-independent forward translocation appear to be strongly stabilized by raising salt into the physiological range and by lowering pH (). The NTP-independent stabilization of the post-translocated TEC is not expected to depend on the protonation state of H936, which likely requires tight trigger loop closing on a cognate NTP. Observing the exo III footprint of the downstream RNAP boundary at a pH of 5.9, 6.9 and 7.9 and at a KCl concentration of 40 and 150 mM, the effects of lowering pH and/or of approaching physiological salt concentration appear strikingly similar. Both treatments strongly stabilize the forward translocation state of the RNAP TEC both in the absence and in the presence of 400 μM CTP. These results are consistent with a role for a protonated histidine residue (i.e., H936) in retention of CTP in the active site. Stabilization of the post-translocated state of the resting RNAP TEC, in the absence of CTP, however, cannot readily be attributed solely to H936, which might only be expected to interact with the NTP in a post-translocated TEC with a closed trigger loop.Citation3,Citation5 Conserved histidines that line the RNA/DNA channel and the downstream DNA/DNA duplex, therefore, may also be involved in regulating RNAP translocation (as discussed further below).
Figure 2. RNAP stalls in the post-translocation register. Forward translocation stability in the absence and presence of NTPs is strongly stimulated by increasing salt and decreasing pH. (A) Exo III mapping of 3′dA9 TECs +/− cognate CTP (400 μM) pH = 5.9, 6.9 and 7.9 and KCl = 40 and 150 mM. The scaffold is as in . (B) Exo III mapping of 3′dA9 TECs +/− cognate GTP or analogs (400 μM). Reactions are shown at KCl = 40, 60, 80, 100, 120 mM (scaffold as in but with a TDS-65 with 34C specifying GTP substrate and complementary 32G NDS-50 at RNA G10). GDP is < 2% contaminated with GTP, which is sufficient to extend A9 to G10 (lanes 8–11). RNA sequences for the two templates are shown.
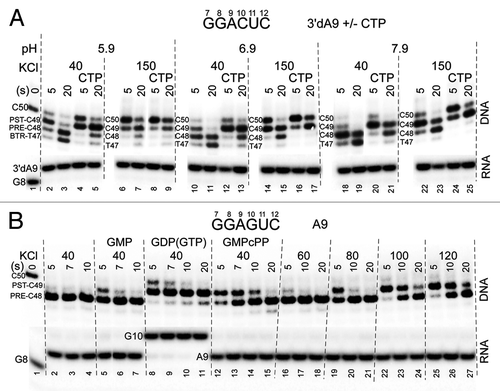
As previously indicated, these data also strongly support the previous model that the RNAP TEC rests in a primarily post-translocated state.Citation1,Citation2 Conditions can be found that stabilize what we interpret as a post-translocated TEC (C50 and C49 DNA bands; exo III digestion to i+19 and i+18),Citation2 in the presence of CTP, even at 20 sec incubation with exo III (; see schematic, ). In the absence of CTP, at 150 mM KCl, the C49 DNA band can still be detected at 20 sec of exo III treatment (). We suggest that the stabilization of the C50 DNA band with CTP (protection at i+19) may be due to strengthened RNAP-DNA contacts near the downstream RNAP boundary at lowered pH and/or at elevated KCl. The C50 band can be observed at higher pH and lower KCl, but, at 40 mM KCl, C50 is much more rapidly attacked by exo III, and the C49 DNA band, therefore, is more strongly detected at earlier time points. If the post-translocated TEC can give rise to the C50 and C49 bands at higher salt and lower pH, the pre-translocated TEC might similarly be resolved as C49 and C48 bands under these conditions (exo III digestion to i+19 and i+18; ). At higher salt and lower pH, therefore, it should be considered whether some of the C49 signal arises from a pre-translocated TEC with enhanced protection of the C49 signal at the downstream RNAP TEC boundary. Higher salt and lower pH enhance the contacts of RNAP to DNA, increasing the duration of protection at i+19 from exo III attack (see schematic, ).
Transcriptional fidelity
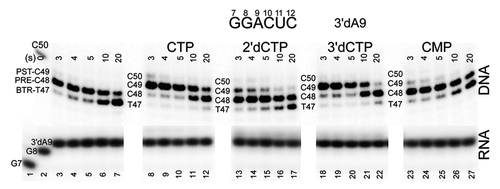
Because an accurately templated NTP loaded to the i+1 site so strongly supports forward translocation for the chain terminated 3′dA9 TEC, NTP analogs can be substituted for a cognate NTP to obtain a ranking for transcriptional fidelity of NTP loading to the RNAP active site. In this analysis, furthermore, analogs can be tested that are not substrates for incorporation. In and , we show a ranking of GTP and CTP (400 μM) analogs for 3′dA9 TECs using appropriate template (TDS-65) and, over their shorter length, fully complementary non-template (NDS-50) DNA strands. For GTP (), at 40 mM KCl, the ranking (most stabilizing of forward translocation to least stabilizing) is GTP~GMPcPP > 2’dGTP > GMP > no NTP~UTP~CTP~ATP. With a 3′dA9 chain terminator, non-cognate NTPs appear to have little or no capacity to stabilize forward translocation. If trigger loop closing were necessary to stabilize the forward position of the TEC, this result with non-cognate NTPs might be expected. At 150 mM KCl, the ranking is GTP > 3′-dGTP > 3′-OCH3-GTP > 2’-dGTP > GDP(GTP at < 2%) > GMP > GMP + 1 mM PPi~no NTP. GDP preparations are contaminated at a low level (< 2%) with GTP (, lanes 8–11, and data not shown). GMP may be contaminated with GDP but not GTP (, lanes 5–7). As shown in , for CTP as the incoming cognate nucleotide, at 40 mM KCl, the ranking is CTP~3′-dCTP > 2’-dCTP > CMP > no NTP. These results are completely consistent with expectations.Citation12,Citation13 Significantly, 3′-dNTPs and other analogs not found abundantly in nature are strongly preferred to stabilize forward translocation over 2’-dNTPs, which are abundant and are the normal substrate for DNA rather than RNA synthesis and should therefore be rejected by RNAPs. Apparently, RNAP is evolved to specifically exclude 2’-dNTPs.Citation12,Citation13 These experiments show that the exo III assay can be used to determine the fidelity of NTP loading to the RNAP active site, even for analogs that are not RNAP substrates, depending on reagent purity. At 400 μM, stabilization of forward translocation using some analogs might be partly attributed to contamination with GTP, but results with analogs are very similar at much lower concentrations (not shown). GMPcPP, GMP, 3′-dGTP and 3′-OCH3-GTP are not substantially GTP contaminated based on their use in our assays and controls.
E. coli RNAP appears to load one NTP at a time
Because the exo III assay shows such robust effects of the incoming NTP, the system was used to detect potential loading of multiple NTPs on template simultaneously.Citation24-Citation32 In Figure S2, the capacity of 25, 100, 200 and 400 μM GTP to stabilize the post-translocated RNAP 3′dA9 TEC was tested using a template encoding a 8-GAGU-11 RNA sequence. A range of GTP concentrations was selected because either an increase or decrease in the stability of the post-translocated TEC induced by NTPs could potentially be observed. If the next template position (i+2) could be occupied with cognate UTP, we reasoned that UTP might increase or decrease the stability of the post-translocated 3′dA9 TEC in the presence of GTP (i+1). In Figure S2, however, no strong effect of addition of UTP (i+2 cognate) or ATP (i+2 non-cognate) was observed. These results appear most consistent with the loading of only a single NTP at or near to i+1 for E. coli RNAP. Because incorporation of GMP is blocked by the 3′dA9 chain terminator, it appears that GMP incorporation must occur prior to UTP loading to template. Furthermore, in this assay, little competition between non-cognate NTPs and cognate GTP at i+1 is observed. If trigger loop closing and H936 protonation are required for stable cognate GTP loading, low competition by non-cognate NTPs is expected, because the closed trigger loop should shield the cognate GTP substrate from exchange.
The 3′ end of the RNA
In , the 3′-end of the RNA (i site for a post-translocated TEC; i+1 site for a pre-translocated TEC) was investigated for its ability to stabilize the forward translocation state. One reason for doing this experiment was to show that exo III patterns at the downstream TEC boundary were specified by the RNAP active site. Another reason was to rank different chain terminators for their effects on the rate of the post- to pre-translocation state transition and their responses to cognate NTPs. As expected, a 3′-OH is slightly more stable for the post-translocated register of the TEC than a 3′-H2 RNA end. A 3′-H2 chain terminator, however, is much more stable for maintenance of the post-translocated state than a bulky 3′-OCH3 RNA end, which is also a chain terminator. Chain terminated ends were tested for GTP and GTPαS loading. Both GTP and GTPαS strongly stabilize forward translocation of the 3′-H2 TEC. Using the bulky 3′-OCH3 RNA end, GTP and GTPαS weakly stimulate forward translocation. Surprisingly, GTPαS at 400 μM appears to load more stably to the active site than GTP using either a 3′-H2 or a 3′-OCH3 chain terminator. This may indicate the utility of GTPαS for some applications using the exo III translocation assay, for instance, varying pH and/or salt or analyzing some RNAP mutant proteins. Similarly, 3′-OCH3 RNA ends may be useful to stabilize the pre-translocated TEC, for instance, in exo III mapping from the RNAP TEC upstream boundary, at which the pre-translocated TEC may be difficult to observe.Citation2 Because alterations to the RNA 3′ end have large and predictable effects on translocation, this demonstrates the specificity of the exo III mapping assay to the RNAP TEC. Large differences in exo III digestion patterns are induced by very subtle chemical changes of the RNA 3′ end, sequestered within the deeply buried RNAP active site, far from the site of exo III digestion (see ). Such changes in exo III patterns cannot be attributed easily to any non-specific effect of exo III activity.
Acid catalysis in NTP loading and translocation
These studies show that the resting translocation register for RNAP at A9 is primarily post-translocated.Citation1,Citation2 Increasing salt and decreasing pH strongly stabilizes the post-translocation register of the RNAP TEC. Salt and pH effects may indicate the involvement of protonated histidine residues, to stabilize forward translocation in both the absence and presence of a cognate NTP,Citation22 as discussed further below. Stabilization of forward RNAP translocation in the presence of NTPs could be attributed to H936 on the trigger loop (, right panel), which is thought to be protonated during cognate NTP loading.Citation9-Citation11,Citation23 In data submitted for review, we show that a H936A substitution has predictable defects in NTP recognition and response to KCl concentration using exo III assays.
In the absence of NTPs, highly conserved histidines lining the nucleic acid binding main channel of RNAP may contribute to pH and salt effects on translocation. β’ H469 near the active site and β H447, H526 (fork region), H1237 and H1244 (switch 3 region) are candidates for histidines that may interact with or closely approach nucleic acids and influence the salt and pH dependence of translocation. Histidine protonation can be important in sequence-specific DNA binding. As examples, human papilloma virus E2 proteinCitation33 and human glucocorticoid receptorCitation34 have been shown to utilize histidine protonation as a mechanism for enhancing sequence-specific DNA recognition. For RNAP translocation and NTP loading, specific histidines are hypothesized to be protonated only when on their DNA, RNA or NTP target. Trigger loop closing on a cognate NTP, therefore, might provide a specific microenvironment for H936 protonation. During translocation, histidines in the main RNAP channel could regulate movement by deprotonating during sliding and reprotonating on the next phosphate downstream. Based on results with RNAP, HPV E2 and glucocorticoid receptor, we suggest that histidine protonation/deprotonation may be underappreciated as a mechanism for enhancing the strength and specificity of protein-nucleic acid interactions.
Stabilization by lowered pH and increased salt appears most consistent with histidine protonation,Citation22 but in an enclosed Zwitterionic environment, lysine and arginine may also contribute to salt and pH effects on RNAP translocation. Arginine does not become deprotonated under physiological conditions.Citation35 Furthermore, solvent exposed lysine, as in the RNAP main channel, is rarely deprotonated.Citation36,Citation37 Opening and closing of downstream DNA at i+2, the RNA/DNA hybrid at i-8 and DNA at i-11 or i-12, associated with the translocation step, is expected to be more strongly affected by KCl concentration than by pH (see ) because nucleic acids do not include easily titrated groups. Charged histidine, arginine, lysine, glutamate and aspartate side chains are stabilized by elevated salt. Here we show surprisingly potent effects on RNAP translocation within the 40–150 mM KCl concentration range. We show effects on translocation in the pH range of 5.9 to 7.9, and histidine(s) appears the most likely target.
Overall, the RNAP mechanism appears to be general base catalyzed because of the strong dependence on deprotonation of the 3′-OH of the RNA (i site; post-translocated TEC) for attack on the α-phosphate of the NTP substrate (i+1 site; post-translocated TEC; ).Citation13 The RNAP mechanism, however, is expected to have an acid catalyzed component, which is the transfer of a proton first from solvent to H936 and then to the β-phosphate of the NTP during phosphodiester bond formation.Citation9-Citation11,Citation23 In this work, we have uncovered acid catalyzed components of RNA synthesis by creating an assay for stabilizing forward translocation that is strongly stimulated by the incoming cognate NTP, lowered pH and/or elevated salt. Somewhat surprisingly, we show that forward translocation in the absence of a NTP substrate also appears to be strongly stabilized by reduced pH and by increased salt. These experiments reveal the translocation resting state of RNAP to be primarily post-translocated and, also, provide a powerful approach to analyze RNAP fidelity and RNAP mutant proteins. These studies also show that analysis of transcription must be multi-dimensional because of the surprisingly strong effects of variables such as pH and salt on RNAP TEC interactions to nucleic acids and substrates. The analysis of the pH dependence of RNA elongation, for instance, was done at 40 mM KCl,Citation13 and, based on the results shown here, should probably be repeated at higher salt concentrations to limit RNAP sliding. Restricting RNAP motion may also be important in analyzing transcriptional fidelity. Based on exo III mapping, any assay that involves RNAP TEC mapping (DNase I and KMnO4 footprinting, for examples) should be done as a function of salt and pH to restrict or increase RNAP sliding. The conditions of 40 mM KCl and pH 7.9 appear to have been chosen originally based on promoter initiation assays and may not be the optimum condition to analyze elongation or RNAP-nucleic acid interactions. It appears that RNAP TECs slip upstream much more rapidly under the 40 mM KCl and pH 7.9 condition compared with the 150 mM KCl and/or pH 5.9 conditions.
Methods
Exo III mapping of the RNAP downstream border was done essentially as previously described.Citation2,Citation17-Citation19 Reactions were done at 21 C. Briefly, histidine-tagged E. coli RNAP was assembled with G7 RNA (5′-CGAGAGG-3′) annealed to TDS-65 template DNA strand (). TDS-65 has a sulfur for oxygen substitution at the 3′ end to block exo III digestion. NDS-50 non-template DNA strand was added. As described in the text, figures and legends, some experiments were done with TDS-65 and a template strand 34C (specifying GTP) and some with a template strand 34G (specifying CTP) at the RNA 10 position (see ). Over their length, NDS-50 DNAs were fully complementary. G7 RNA and NDS-50 were 5′-end labeled with 32P for detection on gels. TECs were immobilized on Promega Magnesphere Ni-beads for bulk preparation. Before assay, TECs were released from beads with 150 mM imidazole, pH 7.0. TECs were then diluted and distributed between sample tubes for other additions, as indicated in the text, figures and legends. The final imidazole concentration after dilution was < 5 mM. No major differences were noted with exo III mapping on beads with no imidazole addition or dissociated from beads with imidazole (data not shown). The exo III reaction buffer was 20 mM TRIS-HCl pH 7.9, 40 to 150 mM KCl (as indicated), 5 mM MgCl2, 5% glycerol, 0.003% IGEPAL CA-630 (Sigma), 10 μM ZnS04 and 1 mM β-mercaptoethanol. Short time points were made more precise using a metronome application on a cell phone to synchronize rapid additions. DNA and RNA products were detected on gels and analyzed with a phosphorimager (Amersham Biosciences; Typhoon 9200).
| Abbreviations: | ||
| exo III | = | exonuclease III |
| GMPcPP | = | α,β-methylene guanosine triphosphate |
| GTPαS | = | guanosine-5′-(α-thio)-triphosphate |
| 3′-OCH3-GTP | = | 3′-O-methyl-GTP |
| NTP | = | nucleoside triphosphate |
| nt | = | nucleotide |
| PPi | = | pyrophosphate (diphosphate) |
| PPiH | = | protonated pyrophosphate |
| RNAP | = | RNA polymerase |
| RNA-A9 | = | 9 nucleotide RNA ending in 3′-AMP |
| TEC | = | ternary elongation complex (RNAP, DNA, nascent RNA) |
Additional material
Download Zip (1.4 MB)Acknowledgments
This work was supported by the National Science Foundation MCB-1050867 (70%) (to ZFB (PI) and Robert I. Cukier (co-PI)) and the National Institutes of Health R01 GM 092949 (30%) to Michael Feig (PI) and ZFB (co-PI). We thank Maria Kireeva for a critical reading of this manuscript. We are very grateful to the Mikhail Kashlev laboratory for invaluable help, support and advice setting up exo III assays. H936A RNAP was from Georgi Belogurov. ZFB receives support from Michigan State University, the Michigan State University Agricultural Experiment Station, and the Michigan State University College of Osteopathic Medicine.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res 2012; 40:7442 - 51; http://dx.doi.org/10.1093/nar/gks383; PMID: 22570421
- Nedialkov YA, Nudler E, Burton ZF. RNA polymerase stalls in a post-translocated register and can hyper-translocate. Transcr 2012; •••:3
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 2006; 127:941 - 54; http://dx.doi.org/10.1016/j.cell.2006.11.023; PMID: 17129781
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 2007; 448:157 - 62; http://dx.doi.org/10.1038/nature05932; PMID: 17581590
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature 2007; 448:163 - 8; http://dx.doi.org/10.1038/nature05931; PMID: 17581591
- Brueckner F, Armache KJ, Cheung A, Damsma GE, Kettenberger H, Lehmann E, et al. Structure-function studies of the RNA polymerase II elongation complex. Acta Crystallogr D Biol Crystallogr 2009; 65:112 - 20; http://dx.doi.org/10.1107/S0907444908039875; PMID: 19171965
- Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol 2008; 15:811 - 8; http://dx.doi.org/10.1038/nsmb.1458; PMID: 18552824
- Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J 2011; 30:4755 - 63; http://dx.doi.org/10.1038/emboj.2011.396; PMID: 22056778
- Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Götte M, et al. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A 2007; 104:4267 - 72; http://dx.doi.org/10.1073/pnas.0608952104; PMID: 17360513
- Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol 2009; 16:212 - 8; http://dx.doi.org/10.1038/nsmb.1540; PMID: 19151724
- Carvalho ATP, Fernandes PA, Ramos MJ. The Catalytic Mechanism of RNA Polymerase II. J Chem Theory Comput 2011; 7:1177 - 88; http://dx.doi.org/10.1021/ct100579w
- Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol 2010; 17:99 - 104; http://dx.doi.org/10.1038/nsmb.1732; PMID: 19966797
- Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, et al. Stepwise mechanism for transcription fidelity. BMC Biol 2010; 8:54; http://dx.doi.org/10.1186/1741-7007-8-54; PMID: 20459653
- Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell 2008; 30:547 - 56; http://dx.doi.org/10.1016/j.molcel.2008.04.023; PMID: 18538653
- Andrecka J, Treutlein B, Arcusa MA, Muschielok A, Lewis R, Cheung AC, et al. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res 2009; 37:5803 - 9; http://dx.doi.org/10.1093/nar/gkp601; PMID: 19620213
- Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci U S A 2004; 101:4776 - 80; http://dx.doi.org/10.1073/pnas.0307241101; PMID: 15037753
- Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell 2008; 30:557 - 66; http://dx.doi.org/10.1016/j.molcel.2008.04.017; PMID: 18538654
- Hein PP, Palangat M, Landick R. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry 2011; 50:7002 - 14; http://dx.doi.org/10.1021/bi200437q; PMID: 21739957
- Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell 2007; 27:406 - 19; http://dx.doi.org/10.1016/j.molcel.2007.06.008; PMID: 17679091
- Nedialkov YA, Opron K, Assaf F, Artsimovitch I, Kireeva ML, Kashlev M, et al. The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation. Biochim Biophys Acta 2013; 1829:187 - 98; http://dx.doi.org/10.1016/j.bbagrm.2012.11.005; PMID: 23202476
- Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J 2012; 31:630 - 9; http://dx.doi.org/10.1038/emboj.2011.432; PMID: 22124324
- Baran KL, Chimenti MS, Schlessman JL, Fitch CA, Herbst KJ, Garcia-Moreno BE. Electrostatic effects in a network of polar and ionizable groups in staphylococcal nuclease. J Mol Biol 2008; 379:1045 - 62; http://dx.doi.org/10.1016/j.jmb.2008.04.021; PMID: 18499123
- Svetlov V, Nudler E. Basic mechanism of transcription by RNA polymerase II. Biochim Biophys Acta 2013; 1829:20 - 8; http://dx.doi.org/10.1016/j.bbagrm.2012.08.009; PMID: 22982365
- Burton ZF, Feig M, Gong XQ, Zhang C, Nedialkov YA, Xiong Y. NTP-driven translocation and regulation of downstream template opening by multi-subunit RNA polymerases. Biochem Cell Biol 2005; 83:486 - 96; http://dx.doi.org/10.1139/o05-059; PMID: 16094452
- Nedialkov YA, Gong XQ, Hovde SL, Yamaguchi Y, Handa H, Geiger JH, et al. NTP-driven translocation by human RNA polymerase II. J Biol Chem 2003; 278:18303 - 12; http://dx.doi.org/10.1074/jbc.M301103200; PMID: 12637520
- Gong XQ, Zhang C, Feig M, Burton ZF. Dynamic error correction and regulation of downstream bubble opening by human RNA polymerase II. Mol Cell 2005; 18:461 - 70; http://dx.doi.org/10.1016/j.molcel.2005.04.011; PMID: 15893729
- Xiong Y, Burton ZF. A tunable ratchet driving human RNA polymerase II translocation adjusted by accurately templated nucleoside triphosphates loaded at downstream sites and by elongation factors. J Biol Chem 2007; 282:36582 - 92; http://dx.doi.org/10.1074/jbc.M707014200; PMID: 17875640
- Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell 2001; 106:243 - 52; http://dx.doi.org/10.1016/S0092-8674(01)00420-2; PMID: 11511351
- Holmes SF, Erie DA. Downstream DNA sequence effects on transcription elongation. Allosteric binding of nucleoside triphosphates facilitates translocation via a ratchet motion. J Biol Chem 2003; 278:35597 - 608; http://dx.doi.org/10.1074/jbc.M304496200; PMID: 12813036
- Kennedy SR, Erie DA. Templated nucleoside triphosphate binding to a noncatalytic site on RNA polymerase regulates transcription. Proc Natl Acad Sci U S A 2011; 108:6079 - 84; http://dx.doi.org/10.1073/pnas.1011274108; PMID: 21447716
- Johnson RS, Strausbauch M, Carraway JK. Rapid pyrophosphate release from transcriptional elongation complexes appears to be coupled to a nucleotide-induced conformational change in E. coli core polymerase. J Mol Biol 2011; 412:849 - 61; http://dx.doi.org/10.1016/j.jmb.2011.05.023; PMID: 21624374
- Johnson RS, Strausbauch M, Cooper R, Register JK. Rapid kinetic analysis of transcription elongation by Escherichia coli RNA polymerase. J Mol Biol 2008; 381:1106 - 13; http://dx.doi.org/10.1016/j.jmb.2008.06.089; PMID: 18638485
- Eliseo T, Sánchez IE, Nadra AD, Dellarole M, Paci M, de Prat Gay G, et al. Indirect DNA readout on the protein side: coupling between histidine protonation, global structural cooperativity, dynamics, and DNA binding of the human papillomavirus type 16 E2C domain. J Mol Biol 2009; 388:327 - 44; http://dx.doi.org/10.1016/j.jmb.2009.03.013; PMID: 19285507
- Lundbäck T, van Den Berg S, Härd T. Sequence-specific DNA binding by the glucocorticoid receptor DNA-binding domain is linked to a salt-dependent histidine protonation. Biochemistry 2000; 39:8909 - 16; http://dx.doi.org/10.1021/bi000231i; PMID: 10913303
- Harms MJ, Schlessman JL, Sue GR, García-Moreno B. Arginine residues at internal positions in a protein are always charged. Proc Natl Acad Sci U S A 2011; 108:18954 - 9; http://dx.doi.org/10.1073/pnas.1104808108; PMID: 22080604
- Harms MJ, Schlessman JL, Chimenti MS, Sue GR, Damjanović A, García-Moreno B. A buried lysine that titrates with a normal pKa: role of conformational flexibility at the protein-water interface as a determinant of pKa values. Protein Sci 2008; 17:833 - 45; http://dx.doi.org/10.1110/ps.073397708; PMID: 18369193
- Isom DG, Castañeda CA, Cannon BR, García-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci U S A 2011; 108:5260 - 5; http://dx.doi.org/10.1073/pnas.1010750108; PMID: 21389271
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996; 14:33 - 8, 27-8; http://dx.doi.org/10.1016/0263-7855(96)00018-5; PMID: 8744570