Abstract
Vibrioparahaemolyticus ExsA is the transcriptional regulator for type III secretion system 1 (T3SS1) while ExsD blocks T3SS1 expression. Herein we show that deletion of exsC from V. parahaemolyticus blocked synthesis of T3SS1-dependent proteins under inducing conditions (contact with HeLa cells), while in trans complementation of the ΔexsC strain with wild-type exsC restored protein synthesis. Under non-inducing conditions (Luria broth plus salt), in trans expression of exsC in a wild-type strain resulted in synthesis and secretion of T3SS1-dependent proteins. Deletion of exsC does not affect the synthesis of ExsA while expression of T3SS1 genes is independent of ExsC in the absence of ExsD. Co-expression of recombinant proteins with different antigenic tags demonstrated that ExsC binds ExsD and that the N-terminal amino acids of ExsC (positions 7 to 12) are required for binding. Co-expression and purification of antigentically tagged ExsA and ExsD demonstrated that ExsD directly binds ExsA and presumably prevents ExsA from binding promoter regions of T3SS1 genes. Collectively these data demonstrate that ExsD binds ExsA to block expression of T3SS1 genes, while ExsC binds ExsD to permit expression of T3SS1 genes. ExsA, ExsC, and ExsD from V. parahaemolyticus appear to be functional orthologues of their Pseudomonasaeruginosa counterparts.
Introduction
Vibrio parahaemolyticus is a Gram-negative marine pathogen that can be transmitted to humans through consumption of contaminated fish and shellfish.Citation1–Citation6 The main manifestation of V. parahaemolyticus infection is gastroenteritis with diarrhea, abdominal pain, vomiting, headache, fever and chills.Citation6V. parahaemolyticus-induced diarrhea is inflammatory with edema, congestion of blood vessels, hemorrhage and increased level of neutrophils in the lamina propria of the intestine.Citation7 In contrast, V. cholerae causes non-inflammatory, secretory diarrhea resulting in severe and rapid dehydration and shock.Citation8 In addition to gastroenteritis, V. parahaemolyticus also causes septicemia, particularly for individuals with preexisting liver disease.Citation9,Citation10
Thermostable direct hemolysin (TDH) is a well-known exogenous toxin that is produced by clinical strains of V. parahaemolyticus.Citation5 Cell culture studies have shown that TDH is required to alter ion flux and to form pores in intestinal cell membranes.Citation11,Citation12 Using a rabbit ileal loop model, Nishibuchi et al.Citation13 demonstrated that fluid accumulation following infection with a tdh deletion strain was significantly reduced compared to the wild-type strain. Other studies showed that the pathogenicity of V. parahaemolyticus was also related to the adherence to human epithelial cells,Citation14 production of TDH-related hemolysin (TRH)Citation15 and vibrioferrin.Citation16 Although our knowledge of V. parahaemolyticus pathogenesis continues to expand, a comprehensive understanding of the molecular mechanisms that are responsible for the inflammatory diarrhea has not been defined.
Type III secretion systems (T3SS) were first discovered in YersiniaCitation17,Citation18 and it has been subsequently shown that many Gram-negative bacteria harbor T3SSs.Citation19 A T3SS is composed of a basal body that spans the periplasmic space, a “needle” that extends from the bacterial membrane surface, and a translocator apparatus connecting the needle with the eukaryotic cell membrane.Citation20–Citation22 This needle-like structure allows bacteria to inject effector proteins directly into host cells where they interfere with normal cell physiology leading to a variety cellular responses.Citation23–Citation25 The genome of V. parahaemolyticus encodes two T3SSs (T3SS1 and T3SS2).Citation26 T3SS1 is responsible for cytotoxicity while T3SS2 appears to contribute to enterotoxicity.Citation27,Citation28 Initial studies showed that T3SS1 of V. parahaemolyticus induced apoptosis.Citation29,Citation30 Later studies using the same strain showed that T3SS1 of V. parahaemolyticus induced autophagyCitation31 but not apoptosis. Inhibition of autophagy does not block T3SS1-induced cytotoxicity against HeLa cellsCitation31 indicating that other mechanisms contribute to T3SS1-induced cell death. Using a different strain (NY-4), we have shown that T3SS1 induces cell-death in HeLa and U937 cells in a manner consistent with oncosis. The interaction is characterized by pore formation in the membrane, presence of active Poly ADP ribose polymerase (PARP) and the process is caspase-independent.Citation27 More importantly, addition of osmoprotectants reduces cytotoxicity against both HeLa and U937 cellsCitation27 indicating that oncosis is, at least partially, the outcome of T3SS1-induced cytotoxicity.
The effector proteins responsible for oncosis have not been identified, although multiple V. parahaemolyticus effector proteins have been identified.Citation29,Citation30,Citation32–Citation36 For example, VopS (Vp1686) is secretedCitation29 and translocatedCitation33,Citation37 into host cells by T3SS1 where it inhibits Rho GTPaseCitation37 and NFkappaB activity,Citation30 leading to depolymerization of actin structureCitation37 and apoptosis,Citation30 respectively. Further studies show that VopS (Vp1686) inhibits Rho GTPase activity by modifying a conserved residue with AMP.Citation36 VopL (Vpa1370) is secreted by T3SS2 of V. parahaemolyticus and it induces formation of long actin stress fibers resulting in the disruption of actin homeostasis.Citation35
Expression of T3SSs in Gram-negative bacteria is controlled by specific environmental conditions that trigger activity from specific transcriptional regulators.Citation38,Citation39 For example, the central regulator, HilA, controls expression of SPI1 in Salmonella by directly binding to the promoters of prg/org and inv/spa operons.Citation40,Citation41 In Yersinia, an AraC-like transcriptional factor, YsaE, regulates expression of Ysa secretion system (sycByspBCDA operon).Citation42 Transcriptional factor, ExsA, is required for the low calcium-induced expression of T3SS in Pseudomonas.Citation43 ExsD binds ExsA directly and this blocks ExsA transcriptional activityCitation44,Citation45 while ExsC binds ExsD to permit ExsA transcriptional activity.Citation43,Citation46
The T3SS1 in V. parahaemolyticus may be regulated in a manner similar to the T3SS in P. aeruginosa. The T3SS1 in V. parahaemolyticus is activated by growing bacteria in Dulbecco's modified Eagle's medium (DMEM) or in contact with eukaryotic cells.Citation47 The ExsA homologue in V. parahaemolyticus (Vp1699) is required for the positive regulation of T3SS1 by binding to a promoter motif that is different from Pseudomonas.Citation47 In trans expression of the exsD homologue in V. parahaemolyticus (Vp1698) blocks expression of T3SS1 genes even under inducing conditions.Citation47 The mechanism by which ExsD inhibits transcription or expression of the T3SS1 is unknown, but if ExsA and ExsD are truly functional orthologues to these proteins in P. aeruginosa, then we hypothesized that ExsD binds ExsA to block transcriptional activity.Citation44,Citation45 Furthermore, we hypothesized that the ExsC homologue in V. parahaemolyticus (Vp1701) interacts with ExsD to permit ExsA transcriptional regulator activity. If this hypothesis is correct, then vp1701 is a functional orthologue of exsC in P. aeruginosa. The results reported herein verify that despite the non-overlapping ecological niches of these organisms and considerable amino acid divergence between ExsA, ExsC and ExsD and their P. aeruginosa counterparts, these proteins in V. parahaemolyticus are functional orthologues of the same proteins in P. aeruginosa.
Results
ExsC is required for the expression of T3SS1 genes.
vp1701 is located proximal to the T3SS1 transcriptional regulator exsA (vp1699) and the protein shares 34% amino acid identity with ExsC found in Pseudomonas.Citation46 ExsC regulates expression of T3SS in Pseudomonas by interacting with ExsD and thus the homologous protein in V. parahaemolyticus may have a similar function. We infected HeLa cells or Caco-2 cells with V. parahaemolyticus strain NY-4 (wild-type) and a ΔexsC strain (exsC deletion mutant) and 4 hr after infection we collected the whole-cell lysates to examine the synthesis of two T3SS1 substrates (Vp1656 and Vp1659). As expected, Vp1656 and Vp1659 were synthesized by NY-4 (, top and middle parts) for each infection experiment (, lanes 1 and 3), but neither protein was synthesized by the ΔexsC strain (, lanes 2 and 4). A chaperone protein, DnaK, was included as a loading control (, lower). These results indicate that for cell culture infections exsC is required for the expression of T3SS1 genes in V. parahaemolyticus.
Complementation of exsC restores wild-type phenotype.
To exclude the possibility of polar effects from the exsC deletion, we expressed exsC in trans from the pMMB207 shuttle plasmid and analyzed cell lysate using western blots. Upon infection with HeLa cells, both Vp1656 (, upper) and Vp1659 (, middle) were synthesized by the complemented ΔexsC strain (, lane 8). Overexpression of exsC also induced synthesis of Vp1656 and Vp1659 when cells were grown in LB-S (, lane 3), which is normally a non-inducing condition for T3SS1 expression (, lane 1). Transformation with an irrelevant expression vector had no affect on Vp1656 or Vp1659 synthesis (, lanes 1, 5 and 6). A western blot for DnaK verified that differences in band intensities for different conditions were not due to unequal protein loading on the gels (, lower).
Expression of exsC in trans in wild-type strain activates secretion of Vp1656 and Vp1659 under non-inducing condition.
To further determine if ExsC can activate the entire T3SS1, we determined if Vp1656 and Vp1659 were secreted into the supernatant by the wild-type strain when transformed with the exsC plasmid and cultured in LB-S. As expected, both Vp1656 (, lane 1, upper) and Vp1659 (, lane 1, middle) were not present in the supernatant of NY-4 strain grown in LB-S. Both Vp1656 and Vp1659 were present in the supernatant of NY-4 strain transformed with a plasmid carrying exsC (, lane 2), but were not present in supernatant of NY-4 strain transformed with a plasmid carrying the bla gene (, lane 3) under LB-S growth condition. As positive controls, both the NY-4 strain overexpressing exsA (, lane 4) and the ΔexsD strain (, lane 5) secreted Vp1656 and Vp1659 into the supernatant. To ensure that the absence of Vp1656 and Vp1659 in the supernatant of NY-4 and NY-4:pbla strains was not due the improper protein precipitation, we examined the presence of DnaK in the supernatant (DnaK is secreted independent of T3SS1 expression; data not shown). The results showed that an equal amount of DnaK was present in the supernatant of these strains (, lower). These results indicate that in trans expression of exsC can not only activate the synthesis of Vp1656 and Vp1659 (), but this permits expression of a functional T3SS1 secretion apparatus.
Deletion of exsC does not reduce the expression of exsA under inducing conditions.
Deletion of exsC inhibits expression of T3SS1 genes under inducing conditions () and previous studies showed that ExsA is a transcriptional factor required for the expression of T3SS1 genes.Citation47 Therefore, we examined exsA expression in wild-type (NY-4) and ΔexsC strains under both inducing (infection) and non-inducing (LB-S) conditions after adding an HA tag at the 3′ terminus of exsA (chromosomal). As expected, ExsA-HA was not detectable in NY-4 strain (western blot, anti-HA) when bacteria were grown in LB-S condition (, lane 1), but ExsA-HA was detected from the NY-4 strain under inducing conditions (, lane 2). An identical expression pattern was evident for the ΔexsC strain (, lanes 3 and 4). This experiment shows that under inducing conditions ExsC is not required for ExsA synthesis.
Expression of T3SS1 genes in a ΔexsD strain is independent of ExsC.
T3SS1 genes are constitutively expressed in a ΔexsD strain even when grown under non-inducing conditions (LB-S) and this constitutive expression requires ExsA.Citation47 When we examined T3SS1 gene expression in a ΔexsD background, we confirmed that ExsC is not required for the expression of T3SS1 genes ( and B). This result verifies that ExsC is not directly required for transcription of T3SS1 genes and that in the absence of ExsD, ExsC has no direct influence on T3SS1 transcription or expression.
Concurrent expression shows that ExsC binds Exsd in vivo.
ExsC is required for the expression of T3SS1 genes in a wild-type strain (), but it is not required for T3SS1 gene expression in an exsD deletion strain (), suggesting that regulation of T3SS1 by ExsC occurs indirectly via interactions with ExsD. Because ExsC binds ExsD in Pseudomonas,Citation46 we employed concurrent expression experiments to determine if a similar interaction occurs in V. parahaemolyticus. ExsC and ExsD were tagged with 6xHis and HA antigens, respectively, and were overexpressed by plasmids pDM31 and pMMB207, respectively, in the NY-4 strain. Western blots using antibody against the 6xHis tag or HA tag confirmed that both ExsC (, lane 1, lower) and ExsD (, lane 1, upper) were present in whole-cell lysate of V. parahaemolyticus. Whole-cell lysates were passed through a Ni+ column, washed extensively, and eluted twice. ExsC-6xHis and ExsD-HA were present in both elution fractions (, lanes 2 and 3) consistent with the hypothesis that ExsD-HA was bound to ExsC-6xHis when eluted from the Ni+ column. To exclude the possibility that ExsD-HA bound non-specifically to the Ni+ column, we expressed ExsD-HA alone (, lane 4) in V. parahaemolyticus and did not detect the HA-tagged protein in the elution fractions (, lanes 5 and 6). These results demonstrate that ExsC binds ExsD in V. parahaemolyticus.
The N-terminus of ExsC is required to bind ExsD.
To identify the region of ExsC that is needed to bind ExsD, we removed 34 amino acids from the N-terminus of ExsC and the truncated ExsC-6xHis protein was expressed together with ExsD-HA in V. parahaemolyticus. Because truncation of ExsC results in a smaller sized protein that can be degraded, we fused the truncated ExsC-6xHis with Glutathione S-transferase (GST) at the N-terminus (; pGST_exsC_Δ5′-His). As a control, full length ExsC-6xHis was also fused with GST (; pGST_exsC_His) and expressed together with ExsD-HA in V. parahaemolyticus. As expected, both GST-exsC-6xHis and ExsD-HA were present in both whole-cell lysate and elution (, lanes 1 and 2). These results indicated that the GST fusion does not prevent ExsC from binding ExsD. N-terminal truncated GST-ExsC-6xHis (GST_exsC_Δ5′-His) was present in both whole-cell lysate and elution (, lanes 3 and 4, upper). ExsD-HA, however, was only present in the whole-cell lysate (, lanes 3 and 4, lower), indicating that N-terminus of ExsC is required to bind with ExsD. Alternatively, deletion of N-terminus disrupted the proper protein folding, leading to the inability of truncated ExsC to bind ExsD.
Binding of ExsC with ExsD is required for the expression of T3SS1 genes.
Because N-terminal truncation of ExsC disrupted binding with ExsD, we determined if this produces a subsequent loss of T3SS1 expression (). As expected, both Vp1656 and Vp1659 were synthesized when GST-ExsC-His was produced in trans in NY-4 (LB-S; , lane 2). In contrast, neither Vp1656 nor Vp1659 were synthesized when the truncated version of ExsC (GST_exsC_Δ5′-His) was produced (, lane 1). A western blot using anti-His antibody confirmed that both full-length ExsC (GST_ExsC_His) and truncated ExsC (GST_exsC_Δ5′-His) fusions were produced in NY-4 strain (, third). DnaK served as a loading control (fourth). RT-PCR analysis showed that transcription of T3SS1 genes in the NY-4:pGST_exsC_His strain was greater than that in the NY-4:pGST_exsC_Δ5′-His when bacteria were grown in LB-S (). SecY, a housekeeping gene, served as a positive control for the RT-PCR experiment.
Amino acids from 7 to 12 in ExsC are required for the interaction of ExsC with ExsD.
To determine which part of N-terminus in ExsC is required to interact with ExsD, we constructed expression vectors for ExsC with only amino acid (Aa) 7 to Aa148 (GST_ExsC_7-148), Aa13 to Aa148 (GST_ExsC_13-148) or Aa27 to Aa148 (GST_ExsC_27-148). Each construct was expressed together with ExsD-HA in the NY-4 strain. All of the truncated ExsC proteins were present in the whole-cell lysates and elution fractions (, upper). ExsD was detected in the whole-cell lysate of all the strains (, lanes 1, 3 and 5, lower). ExsD was only present in the elution of NY-4 strain with GST_ExsC_7-148 (, lane 2, lower), while absent in the elution of NY-4 strain with GST_ExsC_13-148 or GST_ExsC_27-148 (, lanes 4 and 6, lower). Collectively, these results indicate that amino acids from position 7 to 12 are required for ExsC to bind ExsD.
Gst-fused ExsC does not bind ExsA.
Because expression of GST-fused ExsC permits transcription () and expression () of T3SS1 genes in LB-S, we needed to confirm that this does not involve binding between ExsC and ExsA. GST-fused ExsC (, lower) and ExsA (, upper) were present in the whole-cell lysate after growth in LB-S (, lane 1), but only GST-fused ExsC was present in the elution (, lane 2). These results indicate that GST-fused ExsC does not bind ExsA and therefore expression of T3SS1 genes does not involve a direct interaction between ExsC and ExsA.
ExsA binds ExsD.
Pseudomonas ExsD interacts with ExsA to prevent its DNA binding activity. Previous efforts to test in vitro binding between V. parahaemolyticus ExsD and ExsA were negative (data not shown) so for the present analysis we used co-expression and purification experiments to re-examine this potential interaction. His-tagged ExsA (, lower) and HA-tagged ExsD (, upper) were present in both the whole-cell lysate (, lane 1) and elution fractions after passage over a nickel column (, lane 2). These results indicate that ExsA binds ExsD in V. parahaemolyticus and presumably this prevents ExsA from binding T3SS1 promoter regions.
Discussion
In Pseudomonas aeruginosa ExsD negatively regulates expression of T3SS proteins by binding directly to ExsACitation44 and thereby blocking the ability of ExsA to bind the promoter regions of T3SS genes.Citation45,Citation48 Under limited calcium conditions ExsD is bound by another protein, ExsC, and this blocks the ExsD-ExsA interaction and permits transcription of T3SS genes.Citation46 Although the conditions that are permissible for T3SS1 expression in V. parahaemolyticus probably do not mirror those required for T3SS expression in P. aeruginosa, the proximal means of controlling T3SS1 expression is very similar. At the broadest level, ExsA is a positive transcriptional regulator for T3SS1 genesCitation47 () and ExsA transcriptional activity is blocked when ExsD binds to ExsACitation48 (). This “anti-activator” function (sensuCitation44) of ExsD is blocked when ExsC binds ExsD ().
Under non-inducing conditions, exsD is transcribedCitation47 while exsC is not transcribed (data not shown), which would explain the lack of T3SS1 expression in LB-S, but this does not explain why T3SS1 genes are expressed under non-inducing conditions when exsD is deleted. It is possible that exsA is normally transcribed constitutively at a low level that is barely detectable by RT-PCR () and not detectable by western blot (), while expression is enhanced when other T3SS1 genes are expressed (although exsA is not autoregulated; Zhou et al. 2008). Under this scenario deletion of exsD is sufficient to permit upregulation of T3SS1 genes during growth in a non-inducing condition and enhanced transcription of exsA is promoted through a positive feedback with an unrecognized T3SS1 protein. One alternative is that exposure to LB-S media leads to a minimal increase in exsA transcription, but activity of newly synthesized ExsA is easily blocked by constitutive expression and synthesis of ExsD. Another more complicated alternative would involve ExsD serving a dual function of blocking exsA transcription directly or indirectly while simultaneously binding ExsA and blocking ExsA promoter binding activity. We submit that the latter scenario is not plausible because ExsA is clearly expressed in the exsC deletion mutant () where ExsD synthesis is presumably uninhibited but T3SS1 expression is blocked ().
To better characterize the role of ExsC in this process we first demonstrated that deletion of exsC blocks expression of two T3SS1 genes after contact with HeLa or Caco-2 cells () indicating that ExsC is necessary for T3SS1 gene expression in addition to ExsA. Complementation with a wild-type exsC gene restored the ability of the exsC mutant to express T3SS1 genes after contact with HeLa cells indicating that deficiency in T3SS1 gene expression in the exsC mutant is not due to the polar effect of the gene deletion. In addition, expression of exsC in trans in wild-type or exsC mutant strains resulted in the constitutive expression of T3SS1 genes under inducing and non-inducing conditions (), indicating that the ability of ExsC to block ExsD depends on the relative abundance of these two molecules. These results were similar to what is observed when ExsA is overexpressed under inducing or non-inducing conditions.Citation47 ExsA is synthesized for both wild-type and ExsC mutant strains under inducing conditions, indicating that ExsC is not required for the synthesis of ExsA under inducing conditions (). These results indicate that downregulation of T3SS1 in the exsC deletion strain under inducing conditions is not caused directly by downregulation of transcriptional factor, ExsA. This is different from the observations for Pseudomonas, where deletion of ExsC significantly reduced the expression of ExsA.Citation46 It is also possible that ExsC acts as a co-factor of ExsA and in the absence of ExsC, ExsA cannot activate the expression of T3SS1 genes under inducing condition. We did not, however, observe binding between ExsA and ExsC in V. parahaemolyticus (), implying that ExsC is not a direct partner protein of ExsA. We also showed that ExsC is not required for the transcription and expression of T3SS1 in the absence of ExsD (). ExsC is required for the expression of T3SS1 genes in wild-type strain, but not required for expression of T3SS1 gene in exsD mutant strain, indicating that ExsC regulates T3SS1 gene expression by blocking the inhibition mechanism of ExsD. Finally, in Pseudomonas, ExsC binds ExsDCitation46 and we demonstrated that analogous binding occurs for these homologues in V. parahaemolyticus ().
ExsC has been demonstrated to bind ExsD in Pseudomonas, and this positively regulates T3SS1,Citation46 but there is no experimental data describing which region of the protein is required for this interaction and if this interaction is required for the expression of T3SS. By substitution of charged residues with alanine or glycine, Lykken et al.Citation49 identified several ExsC mutants that were not able to complement the phenotype of exsC mutant. Nevertheless, these mutants were still able to bind ExsD, suggesting that binding of ExsC with ExsD is not sufficient for the induction of T3SS in Pseudomonas. Through a two-part experiment we showed that for V. parahaemolyticus the 34 amino acids at the N-terminus of ExsC are required for the interaction with ExsD () and that amino acids 7–12 are critical to this interaction (), although it remains to be determined if the interaction occurs at these amino acids or if these amino acids are required to form the tertiary structure needed for ExsC to bind ExsD. Our studies also showed that loss of interaction between ExsC and ExsD resulted in the loss of T3SS1 transcription and expression (), indicating that upregulation of T3SS1 by ExsC () is due to the interaction between ExsC and ExsD.
While our studies show that the ExsA-ExsC-ExsD interaction is very similar for V. parahaemolyticus and P. aeruginosa, we also noted several distinctions. Dasgupta et al.Citation46 observed diminished ExsA synthesis for their ΔexsC strain (their Fig. 1E, lanes 7 and 8) whereas there is no evidence that deletion of exsC from V. parahaemolyticus influences ExsA synthesis (). McCaw et al.Citation44 found that while deletion of exsD resulted in transcription of the T3SS regardless of culture conditions, secretion of TTSS proteins still required Ca2+ chelation. In contrast, deletion of exsD from V. parahaemolyticus permits both transcription and secretion of T3SS1-dependent proteins regardless of the culture conditions tested ().Citation47 Finally, ExsA in V. parahaemolyticus recognizes a promoter binding motif that is distinct from the one recognized by ExsA from P. aeruginosa.Citation43,Citation50,Citation51 Dasgupta et al.Citation46 suggested a regulatory model whereby ExsC is normally sequestered by another T3SS protein (ExsE) and thus not available for binding ExsD. Under this scenario, when the T3SS is activated ExsC acts as a chaperone to deliver the sequestering protein to the T3SS apparatus thereby becoming available to bind ExsD. We do not have any data to support or refute this model for V. parahaemolyticus, although a more parsimonious model would only require ExsC to be synthesized when appropriate environmental conditions are encountered.
Regardless of the differences noted above, it is quite remarkable that such phylogenetically distinct (different order, family and genus) and ecologically divergent (halophilic marine organism versus a generalist) species express barely recognizable homologous proteins that essentially retain conserved functions. Between V. parahaemolyticus and P. aeruginosa, the amino acid similarity for ExsA, ExsC and ExsD is only 45%, 34% and 22%, respectively. If these T3SSs were present in the original common ancestor then these functional traits have been conserved >500 million yearsCitation52 for what might otherwise be considered an accessory pathogenicity island in these two facultative pathogens. That is a strong indication that evolutionary pressures have favored retention of the T3SS function within the diverse ecological niches that are occupied by these two organisms.
Materials and Methods
Bacterial strains, plasmids and growth conditions.
V. parahaemolyticus was routinely grown in Luria-Bertani (LB) medium supplemented with 2.5% of NaCl (LB-S) at 37°C with shaking. Escherichia coli S17-1 lambda pir strain was used for gene cloning and construction of gene deletion mutants and was grown in LB at 37°C with shaking. Plasmid pMMB207 was used for the complementation and protein expression. Plasmid pDM4 was used for generate gene deletion mutants in V. parahaemolyticus. Plasmid pDM31 and pRY107 were used in the protein-protein interaction experiment. All of the wild-type and derivative strains of V. parahaemolyticus and E. coli are listed in . Antibiotics were used in the following concentration: 100 µg/ml ampicillin, 50 µg/ml kanamycin, 17 µg/ml chloramphenicol for E. coli, and 5 µg/ml chloramphenicol for V. parahaemolyticus.
Generation of exsC deletion in wild-type and ΔexsD background strains.
Deletion of exsC gene was achieved by homologous recombination as described in previous studies.Citation27,Citation47 Briefly, a 697 bp DNA fragment in the immediate upstream of exsC (vp1701) and a 700 bp DNA fragment in the immediate downstream of exsC were amplified by PCR using primers pairs ExsC_1F/ExsC_1R and ExsC_2F/ExsC_2R, respectively (). The DNA fragment amplified by ExsC_1F/ExsC_1R was digested with restriction enzymes XhoI and XbaI. DNA fragments amplified by ExsC_2F/ExsC_2R were digested with XbaI and BglII. These two digested fragments were ligated with plasmid pDM4 that was digested with XhoI and BglII resulting in the plasmid pDM4_exsC1 + 2. Plasmid pDM4_exsC1 + 2 was electroporated into E. coli S17 resulting in the strain S17_pDM4_exsC1 + 2. Plasmid pDM4_exsC1 + 2 was subsequently transferred from S17 into the wild-type strain of V. parahaemolyticus (NY-4) by conjugation. Chloramphenicol- and ampicillin-resistant transconjugants were selected and inoculated into LB-S broth supplemented with 10% sucrose to facilitate the excision of the plasmid from the chromosome of V. parahaemolyticus. V. parahaemolyticus in LB-S supplemented with sucrose was streaked onto LB-S plates with 10% sucrose and individual colonies were picked and spotted onto LB-S agar and LB-S agar with chloramphenicol. Chloramphenicol-sensitive colonies were picked and grown in LB-S broth for confirmation of exsC deletion by using PCR with primers ExsC_Forward and ExsC_Reverse. One clone with a PCR confirmed exsC deletion was designated as ΔexsC. To generate the exsC gene deletion in ΔexsD strain, plasmid pDM4_exsC1 + 2 was transferred from the S17 strain into the ΔexsD strain by conjugation. Selection, screening and confirmation of exsC deletion were similar to that described above. Mutant strains were designated as ΔexsD exsC.
Complementation of ΔexsC strain with a wild-type exsC gene or an unrelated bla gene.
Full-length exsC gene was amplified by PCR using ExsC_up and ExsC_down as primers and genomic DNA of NY-4 strain as template. PCR product was purified and digested with EcoRI and XbaI before ligation into pMMB207 that was digested with the same enzymes resulting in the plasmid pMMB207_exsC. This plasmid was electroporated into E. coli S17 resulting in the strain S17_pMMB207_exsC. The plasmid was subsequently transferred from E. coli strain into NY-4 and ΔexsC strains of V. parahaemolyticus resulting in ΔexsC_pMMB207_exsC and NY-4_pMMB207_exsC, respectively. A partial bla gene (∼680 bp) was amplified by using Blaup and Bladown as primers and pCX340,Citation53 as template. PCR product for bla-TEM-1 was purified and digested with XbaI and HindIII before being ligated into pMMB207 that was digested with the same enzymes, leading to the plasmid pMMB207_bla. This plasmid was electroporated into E. coli S17 (S17_pMMB207_bla) and subsequently transferred into NY-4 and ΔexsC strains of V. parahaemolyticus resulting in the ΔexsC_pMMB207_bla and NY-4_pMMB207_bla, respectively.
Generation of HA tagged ExsA in the chromosome.
To add an HA tag at the 3′ end of the exsA gene in the chromosome, primers ExsA_HA_Insert_Fwd and ExsA_HA_Insertion_Rev were used to amplify the entire exsA gene with an HA tag at the C-terminus. PCR products were digested with XhoI and XbaI and ligated into the pDM4 plasmid that was digested with the same enzymes, resulting in the plasmid pDM4_exsA_HA_Insertion. This plasmid was electroporated into E. coli S17 resulting in the strain S17_ pDM4_exsA_HA_Insertion. Plasmid pDM4_exsA_HA_Insertion was conjugated into NY-4 and ΔexsC strains resulting in the strain NY-4_pDM4_exsA_HA_Insertion and ΔexsC_pDM4_exsA_HA_Insertion.
Western blot analysis to detect Vp1656 and Vp1659.
For preparation of protein samples from V. parahaemolyticus grown in non-inducing conditions, overnight bacterial culture was diluted in LB-S (1:100) supplemented with antibiotics as needed. Diluted culture was grown in 37°C with aeration for 4 h and pellets were collected by centrifugation for 10 min (4,000 xg). Pellets were resuspended in 1X Phosphate Buffered Saline (PBS) and sonicated until the bacterial resuspension was clear. For preparation of proteins from V. parahaemolyticus-infected host cells, monolayers of HeLa or Caco-2 cells were seeded in 6-well plates and inoculated with overnight bacterial culture to achieve an M.O.I of 100. Four hours after incubation (37°C with 5% of CO2), adherent and non-adherent bacteria were collected by scraping off the HeLa or Caco-2 cells from the plate and centrifugation. Bacteria and HeLa or Caco-2 cells pellets were resuspended in 1X PBS and sonicated until the resuspension became clear. Protein samples were mixed with equal volume of 2X Laemmli buffer before being loaded onto 12% of SDS-PAGE gel. Electrophoresis was carried out for ∼1 h with at 120 V. Separated proteins were transferred from the gel onto the nitrocellulose membrane (Bio-Rad, Hercules, CA). The membrane was blocked with 5% non-fat milk (Bio-Rad) in PBST (PBS containing 0.1% of Tween-20) for 1 h and then probed with primary polyclonal antibodies against Vp1656,Citation47 and Vp1659 (Call et al. unpub. data) in PBST (1:1,000) for 1 h. After being extensively washed, the membrane was probed with secondary goat anti-mouse IgG antibody conjugated to horseradish peroxidase (CalBiochem, San Diego, CA). The blots were developed as described previously.Citation47
RT-PCR analysis.
RT-PCR analysis was performed as described previously.Citation47
Secretion of Vp1656 and Vp1659 by V. parahaemolyticus.
Preparation of secreted proteins in the supernatant was described previously.Citation47 Briefly, supernatant of the bacterial culture was concentrated by addition of trichloroacetic acid (TCA) to achieve a final 10% concentration followed by centrifugation. Pellets were resuspended in acetone and centrifuged again to obtain protein samples. Protein samples were resuspended in 1X Laemmli buffer before being loaded onto SDS-PAGE gels. Presence of Vp1656 and Vp1659 in the supernatant of bacterial culture was determined by western blot as described above.
Generation V. parahaemolyticus strains for co-expression of exsC, exsA and exsD.
To generate a strain that co-expresses exsC and exsD, we cloned exsC and as an IPTG-inducible promoter Ptac into a plasmid, pDM31. The full-length exsC gene (447 bp) was amplified using ExsC_IP_F and ExsC_IP_R () as primers and genomic DNA of NY-4 as template. The Ptac promoter was amplified using Ptac_F and Ptac_R () as primers and plasmid pMMB207 as template. The DNA fragment of Ptac was digested with SacI and SmaI. The DNA fragment of exsC gene was digested with SmaI and SpeI. Digested fragments of exsC and Ptac were ligated into pDM31,Citation54 that was pre-digested with SacI and SpeI, resulting in the plasmid pDM31_Ptac_exsC_His. This plasmid was electroporated into E. coli S17 resulting in the strain S17_ pDM31_Ptac_exsC. The full-length exsD gene was amplified using ExsD_up and ExsD_HA_down as primers () with genomic DNA of NY-4 as template. The amplified DNA fragment of exsD gene was digested with EcoRI and XbaI. Digested fragments were ligated into plasmid pMMB207 that was pre-digested with the same enzymes, resulting in the plasmid pMMB207_exsD_HA. This plasmid was electroporated into E. coli S17 resulting in the strain S17_ pMMB207_exsD_HA. The full-length exsA gene was amplified using ExsA_up and ExsA_HA_down as primers and genomic DNA of NY-4 strain as template. The amplified DNA fragment of exsA was digested with BamHI and XbaI. Digested fragments were ligated into plasmid pMMB207 that was pre-digested with the same enzymes, resulting in the plasmid pMMB207_exsA_HA. This plasmid was electroporated into E. coli S17 resulting in the strain S17_ pMMB207_exsA_HA. His-tagged ExsA was constructed by using primers ExsA_IP_F and ExsA_IP_R, resulting in the plasmid pDM31_Ptac_exsA_His. To determine the interaction between ExsC and ExsD, plasmids pDM31_Ptac_exsC_His and pMMB207_exsD_HA were transferred from corresponding E. coli S17 strains into NY-4 by conjugation, resulting in the strain NY-4:pexsD-HA pexsC-His. Plasmid pMMB207_exsD_HA was transferred into NY-4 by conjugation resulting in the strain NY-4:pexsD-HA.
To generate GST-fused ExsC, a Glutathione S-transferase (GST) gene as well as Ptac prmoter was amplified using GST_FW and GST_RE as primers and plasmid pGEX-KGCitation55 as template. The full-length exsC gene was amplified using GST_ExsC_FW and GST_ExsC_RE as primers and genomic DNA of NY-4 as template. The amplified GST fragment was digested with SacI and SmaI. The DNA fragment of exsC was digested with SmaI and SpeI. Digested fragments of GST and exsC were ligated into pRY107,Citation56 that was pre-digested with SacI and SpeI, resulting in the plasmid pRY107_GST_exsC_His. This plasmid was electroporated into E. coli S17 resulting in the strain S17_pGST_exsC_His. To determine the protein interaction between GST-fused ExsC and ExsD, plasmids pRY107_GST_exsC_His and pMMB207_exsD_HA were transferred from corresponding E. coli S17 strains into NY-4 strain by conjugation resulting in the strain NY-4:pGST-exsC-His pexsD-HA. To determine the protein interaction between GST-fused ExsC and ExsA, plasmids pRY107_GST_exsC_His and pMMB207_exsA_HA were transferred from corresponding E. coli S17 strains into NY-4 by conjugation resulting in the strain NY-4:pGST-exsC-His-pexsA-HA. To determine the protein interaction between ExsD and ExsA, plasmid pDM31_Ptac_exsA_His and pMMB207_exsD_HA were transferred from corresponding E. coli S17 strains into NY-4 by conjugation resulting in the strain NY-4:pexsD-HA-pexsA-His. To determine the protein interaction between the truncated ExsC and ExsD, the GST gene was amplified and digested as described above. A truncated exsC fragment with deletion of 102 bp at the N-terminus was amplified using ExsC_IP_C_FW and ExsC_IP_R as primers and genomic DNA from NY-4 as template. Amplified truncated exsC gene was digested with SmaI and SpeI. Digested GST and truncated exsC were ligated into plasmid pRY107, leading to the plasmid pRY107_GST_ exsC_Δ5′-His. This plasmid was subsequently electroporated into E. coli S17, leading to the strain S17_pGST_ exsC_Δ5′-His. To determine the protein interaction between N-terminal truncated ExsC with ExsD, plasmids pRY107_GST_ exsC_Δ5′-His and pMMB207_exsD_HA were transferred into NY-4 by conjugation resulting in the strain NY-4:pGST_exsC_Δ5′-His-pexsD-HA. To generate a serial truncation of ExsC fused with GST, the exsC gene from Aa7 to Aa148 was amplified using GST_ExsC_C1_FW and GST_ExsC_RE as primers and genomic DNA of NY-4 as template. The amplified GST fragment was digested with SacI and SmaI. The DNA fragment of exsC from Aa7 to Aa148 was digested with SmaI and SpeI. Digested fragments of GST and exsC were ligated into pRY107 that was pre-digested with SacI and SpeI resulting in the plasmid pRY107_GST_exsC (7-148)_His. ExsC from Aa13-148 was amplified using GST_ExsC_C2_FW and GST_ExsC_RE as primers and genomic DNA of NY-4 as template. The amplified GST DNA was digested with SacI and SmaI. The DNA fragment of exsC from Aa13 to Aa148 was digested with SmaI and SpeI. Digested fragments of GST and exsC were ligated into pRY107 that was pre-digested with SacI and SpeI resulting in the plasmid pRY107_GST_exsC (13-148)_His. ExsC from Aa27-148 was amplified using GST_ExsC_C4_FW and GST_ExsC_RE as primers and genomic DNA of NY-4 as template. The amplified GST DNA fragment was digested with SacI and SmaI. The DNA fragment of exsC from Aa27 to Aa148 was digested with SmaI and SpeI. Digested fragments of GST and exsC were ligated into pRY107 that was pre-digested with SacI and SpeI resulting in the plasmid pRY107_GST_exsC (27-148)_His. pRY107_GST_exsC (7-148) and pMMB207_exsD_HA were transformed into NY-4, resulting in the strain NY-4:pRY107_GST_exsC (7-148) pexsD_HA. pRY107_GST_exsC (13-148) and pMMB207_exsD_HA were transformed into NY-4, resulting in the strain NY-4:pRY107_GST_exsC (13-148) pexsD_HA. pRY107_GST_exsC (27-148) and pMMB207_exsD_HA were transformed into NY-4 resulting in the strain NY-4: pRY107_GST_exsC (27-148) pexsD_HA.
Protein interaction assayed by co-purification.
To determine protein interaction between ExsC and ExsD, overnight culture of V. parahaemolyticus strain NY-4:pexsD-HA pexsC-His was diluted (1:100) into LB-S and grown in 37°C for ∼2 h until the OD600 reached ∼0.5. IPTG was added to reach a final concentration of 1 mM and protein expression was induced for an additional 6 h. Bacterial culture was centrifuged for 10 min (4,000 xg) to collect pellets that were resuspended in 1X purification buffer (50 mM NaH2PO4, pH 8.0, 0.5 M NaCl) for sonication. A portion of sonicated protein sample was collected as whole-cell lysate and the rest of the sample was loaded onto an Ni2+ resin column (Invitrogen, Carlsbad, CA). After binding overnight, the column was washed with washing buffer (1X purification buffer supplemented with 30 mM immidazole) eight times. Proteins were subsequently eluted with 0.5 ml of elution buffer (1X purification buffer supplemented with 300 mM immidazole) twice and subsequently designated as Elution 1 and Elution 2, respectively. Western blot analysis was performed to detect the presence of His-and HA-tagged protein in the whole-cell lysates and in the elutions using monoclonal anti-His antibody (Invitrogen) and polyclonal anti-HA antibody. To exclude the possibility that HA-tagged proteins bound the resin column, control strain NY-4:pexsD-HA was grown and protein samples were prepared as described above. Whole-cell lysate and elutions were examined for the presence of His-and HA-tagged proteins. NY-4:pGST-exsC-His pexsD-HA was used to determine the interaction between GST-fused ExsC and ExsD. NY-4:pGST-exsC-His pexsA-HA was used to determine the interaction between GST-fused ExsC and ExsA. NY-4:pexsD-HA-pexsA-His was used to determine the interaction between ExsA and ExsD. NY-4:pGST_exsC_Δ5′-His pexsD-HA was used to determine the interaction between N-terminal truncated ExsC and ExsD. Strain NY-4:pGST-exsC (7-148)_His pexsD-HA was used to determine the interaction between GST-fused ExsC (7-148) and ExsD. Strain NY-4:pGST-exsC (13-148)_His pexsD-HA was used to determine the interaction between GST-fused ExsC (13-148) and ExsD. Strain NY-4:pGST-exsC (27-148)_His pexsD-HA was used to determine the interaction between GST-fused ExsC (27-148) and ExsD.
Figures and Tables
Figure 1 ExsC is required for the synthesis of Vp1656 and Vp1659 upon V. parahaemolyticus infection of HeLa or Caco-2 cells. Wild-type (NY-4) (lanes 1 and 3) and ΔexsC (lanes 2 and 4) strains were used to infect HeLa (Lanes 1 and 2) and Caco-2 (lanes 3 and 4) cells for 4 h. The whole-cell lysates of the infected samples were probed with polyclonal antibody against Vp1656 (upper), Vp1659 (middle) and for the DNA loading control DnaK (lower).
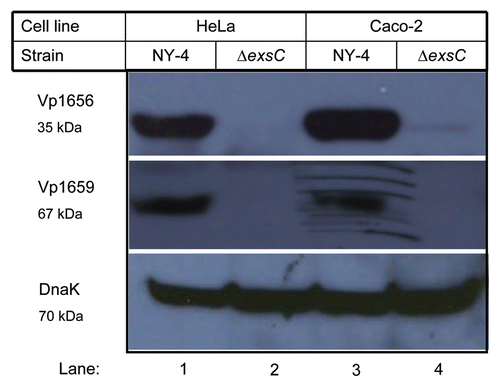
Figure 2 Western blot of whole-cell lysates showing that in trans expression of exsC activates production of Vp1656 and Vp1659. The NY-4 and ΔexsC strains were transformed with an ExsC expression plasmid or control plasmid (pMMB208-bla) and were then grown in non-inducing (LB-S) or inducing conditions (HeLa cell infection) (4 hr). Analysis of the whole-cell lysate showed that NY-4 did not synthesize Vp1656 or Vp1659 when grown in LB (Lane 1) unless ExsC was expressed in trans (Lane 3). In trans expression of exsC led to higher production of both Vp1656 and Vp1659 (Lane 4) compared to wild-type expression levels (Lane 2). A similar pattern of expression (Lanes 5–8) was evident with in trans expression of ExsC for the ΔexsC strain.
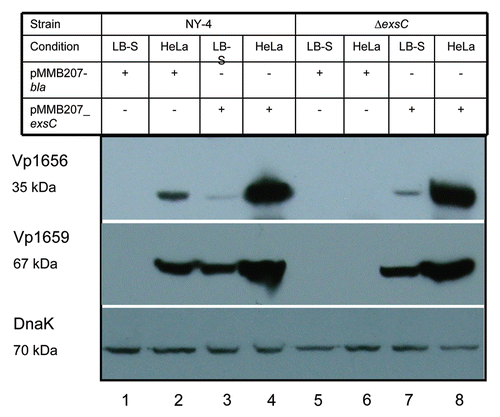
Figure 3 Analysis of secreted proteins shows that ExsC activates secretion of Vp1656 and Vp1659 when grown in non-inducing conditions (LB-S). Proteins were precipitated from the supernatant of NY-4 (lane 1), NY-4:pMMB207_exsC (lane 2), NY-4:pMMB207_bla (lane 3), NY-4:pexsA (lane 4) and ΔexsD (lane 5) strains and probed with polyclonal antibody against Vp1656 (upper), Vp1659 (middle) and DnaK (lower).
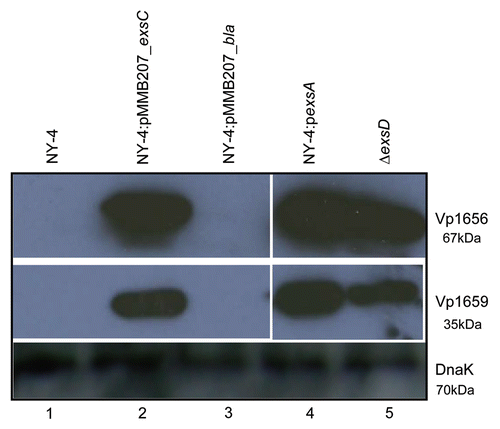
Figure 4 Deletion of ExsC does not reduce the synthesis of ExsA under inducing conditions. For these experiments exsA (chromosomal) was tagged with the HA antigen sequence in both the NY-4 (lanes 1 and 2) and ΔexsC (lanes 3 and 4) strains. After growth in LB-S (lanes 1 and 3) for 4 h or with HeLa cells (lanes 2 and 4) for 4 h, whole-cell lysates were collected and probed with anti-HA (upper) or anti-DnaK (lower) by western blot analysis.
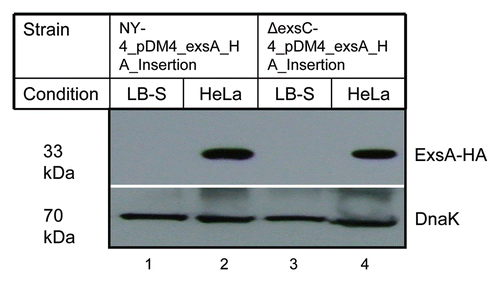
Figure 5 ExsC is not required for the expression of T3SS1 genes in the ΔexsD strain. (A) Protein samples isolated from ΔexsD exsC (lane 1) and ΔexsD (lane 2) strains grown in LB-S were probed with polyclonal antibody against Vp1656 (upper), Vp1659 (middle) and DnaK (lower); (B) RT-PCR analysis was used to detect mRNA transcripts from a subset of T3SS1 genes for the ΔexsD exsC (lane 1) and ΔexsD strains grown in LB-S.
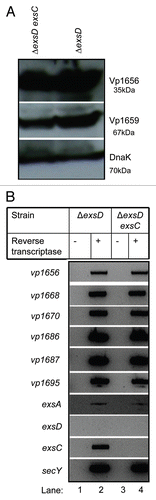
Figure 6 ExsC binds ExsD when these proteins are expressed concurrently. Whole-cell lysates from NY-4:pexsD-HA-pexsC-His (lane 1), NY-4:pexsD-HA (lane 4) were probed with anti-HA (upper; ExsD) and anti-His (lower; ExsC). After passage through a nickel column (which binds the His tag) and washing, eluted fractions from NY-4:pexsD-HA-pexsC-His (lanes 2 and 3), NY-4:pexsD-HA (lanes 5 and 6) were probed with anti-HA (upper) and anti-His (lower) antibodies. ExsC and ExsD were eluted together (lanes 2 and 3) and this outcome was not the result of non-specific binding of the HA protein and the nickel column (lanes 5 and 6).
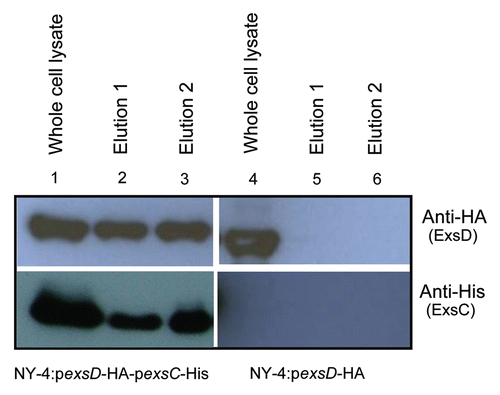
Figure 7 The N-terminus of ExsC is required for ExsC to bind with ExsD. (A) Schematic representation of fusion proteins between full-length exsC and GST (upper), and between N-terminus truncated exsC and GST (lower). “Deletion” indicates the location of the 5-prime deletion for the truncated exsC sequence. (B) Whole-cell lysate and elution from NY-4:pGST-exsC-His pexsD-HA or from NY-4:pGST_exsC_Δ5′-His pexsD-HA strains probed with anti-His (upper) or anti-HA (lower) antibodies. The truncated ExsC_Δ5′protein does not bind ExsD.
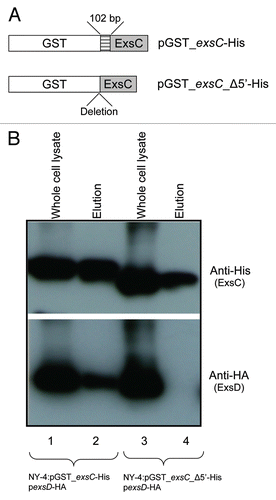
Figure 8 Full-length ExsC is required for transcription and expression of T3SS1 genes. (A) Strains were cultured separately in LB-S and then cell lysates from NY-4:pGST_exsC-_Δ5′-His (lane 1) and NY-4:pGST-exsC-His (lane 2) were probed with anti-Vp1656, anti-Vp1659, anti-His and anti-DnaK; (B) RT-PCR analysis showing mRNA transcripts for a panel of T3SS1 genes for the NY-4:pGST-exsC-His and NY-4:pGST_exsC_Δ5′-His strains after growth in LB-S. SecY, a housekeeping gene, served as a positive control for the RT-PCR experiment.
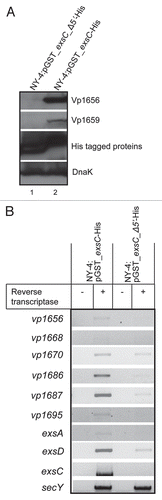
Figure 9 Amino acids from position 7 to 12 of ExsC are required for ExsC to interact with ExsD. Whole-cell lysate and eluted fractions from NY-4:pGST-exsC(7-148)-His pexsD-HA, NY-4:pGST-exsC(13-148)-His pexsD-HA or NY-4:pGST-exsC (27-148)-His pexsD-HA strains were probed with anti-HA (lower) and anti-His (upper) antibodies. “W” indicates whole-cell lysates and “E” indicates eluted fraction.
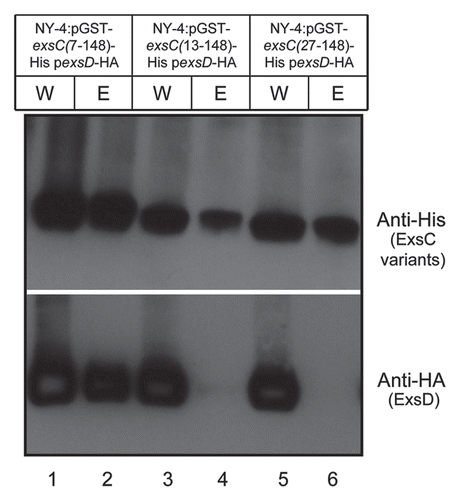
Figure 10 ExsD binds ExsA when expressed concurrently in vivo while GST-fused ExsC does not bind ExsA. Western blots of NY-4 grown in LB-S with co-expression of ExsA with either (A) GST_ExsC_6xHis, or (B) ExsD_HA. ExsA_HA was not eluted with GST_ExsC_6xHis, but ExsD_HA was eluted with ExsA-6xHis indicating that ExsC does not bind ExsA while ExsD does bind ExsA. Specificity of the nickel column for isolation of 6xHis tagged proteins is shown in .
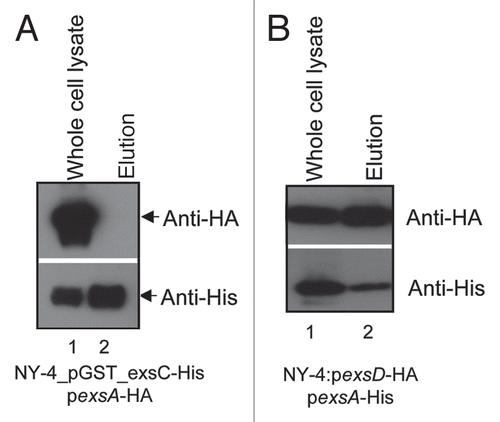
Figure 11 A model illustrating the regulation of T3SS1 genes. When ExsD is present in molar excess of ExsC, ExsD binds ExsA and blocks transcriptional promoter activity. T3SS1 genes are expressed when ExsC binds ExsD or if ExsA is synthesized to a molar concentration in excess of the ExsD concentration. The induction signal for ExsC has not been identified and there is no direct evidence whether or not the same or independent induction signal is required to express and synthesize ExsA (the latter can occur independently of ExsC; ).
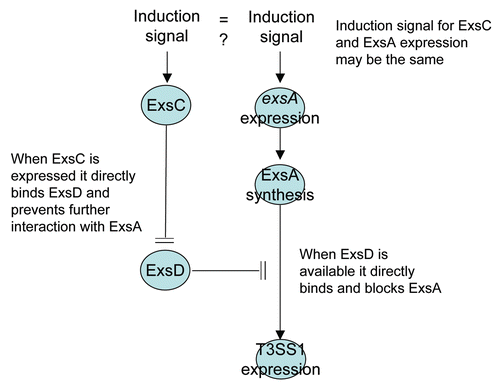
Table 1 Strains and plasmids used in this study
Table 2 Primers used in this study
Acknowledgements
We gratefully acknowledge Lisa Orfe, Patrick Friel, Daniel Erwin, Seth Nydam and Pablo Piñeyro for their technical assistance and discussions. Dr. Kathryn J. Boor provided the wild-type strain of V. parahaemolyticus (NY-4). This project was supported in part by National Institute of Health, Department of Health and Human Services under the contract number NO1-AI-30055 and by the Agricultural Animal Health Program, College of Veterinary Medicine, Washington State University.
References
- DePaola A, Kaysner CA, Bowers J, Cook DW. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas and New York (1997 and 1998). Appl Environ Microbiol 2000; 66:4649 - 4654
- Martinez-Urtaza J, Lozano-Leon A, DePaola A, Ishibashi M, Shimada K, Nishibuchi M, et al. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J Clin Microbiol 2004; 42:4672 - 4678
- Hedberg C. Food-related illness and death in the United States. Emerg Infect Dis 1999; 5:840 - 842
- Mead PS, Slutsker L, Griffin PM, Tauxe RV. Food-related illness and death in the united states reply to Dr. Hedberg. Emerg Infect Dis 1999; 5:841 - 842
- Wong HC, Liu SH, Ku LW, Lee IY, Wang TK, Lee YS, et al. Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J Food Prot 2000; 63:900 - 906
- Yeung PS, Boor KJ. Epidemiology, pathogenesis and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog Dis 2004; 1:74 - 88
- Qadri F, Alam MS, Nishibuchi M, Rahman T, Alam NH, Chisti J, et al. Adaptive and inflammatory immune responses in patients infected with strains of Vibrio parahaemolyticus. J Infect Dis 2003; 187:1085 - 1096
- Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet 2004; 363:223 - 233
- Hlady WG, Klontz KC. The epidemiology of Vibrio infections in Florida, 1981–1993. J Infect Dis 1996; 173:1176 - 1183
- Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis 2000; 181:1661 - 1666
- Honda T, Ni Y, Miwatani T, Adachi T, Kim J. The thermostable direct hemolysin of Vibrio parahaemolyticus is a pore-forming toxin. Can J Microbiol 1992; 38:1175 - 1180
- Takahashi A, Kenjyo N, Imura K, Myonsun Y, Honda T. Cl(−) secretion in colonic epithelial cells induced by the Vibrio parahaemolyticus hemolytic toxin related to thermostable direct hemolysin. Infect Immun 2000; 68:5435 - 5438
- Nishibuchi M, Fasano A, Russell RG, Kaper JB. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun 1992; 60:3539 - 3545
- Hackney CR, Kleeman EG, Ray B, Speck ML. Adherence as a method of differentiating virulent and avirulent strains of Vibrio parahaemolyticus. Appl Environ Microbiol 1980; 40:652 - 658
- Xu M, Yamamoto K, Honda T, Ming X. Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh). J Bacteriol 1994; 176:4757 - 4760
- Wong HC, Lee YS. Regulation of iron on bacterial growth and production of thermostable direct hemolysin by Vibrio parahaemolyticus in intraperitoneal infected mice. Microbiol Immunol 1994; 38:367 - 371
- Michiels T, Wattiau P, Brasseur R, Ruysschaert JM, Cornelis G. Secretion of Yop proteins by Yersiniae. Infect Immun 1990; 58:2840 - 2849
- Salmond GP, Reeves PJ. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci 1993; 18:7 - 12
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 1998; 62:379 - 433
- Galan JE. Energizing type III secretion machines: what is the fuel?. Nat Struct Mol Biol 2008; 15:127 - 128
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 2006; 444:567 - 573
- Galan JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 1999; 284:1322 - 1328
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 2009; 12:117 - 124
- Shao F. Biochemical functions of Yersinia type III effectors. Curr Opin Microbiol 2008; 11:21 - 29
- Backert S, Selbach M. Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol 2005; 13:476 - 484
- Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 2003; 361:743 - 749
- Zhou X, Konkel ME, Call DR. Type III secretion system 1 of Vibrio parahaemolyticus induces oncosis in both epithelial and monocytic cell lines. Microbiology 2009; 155:837 - 851
- Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, et al. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun 2004; 72:6659 - 6665
- Ono T, Park KS, Ueta M, Iida T, Honda T. Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect Immun 2006; 74:1032 - 1042
- Bhattacharjee RN, Park KS, Kumagai Y, Okada K, Yamamoto M, Uematsu S, et al. VP1686, a Vibrio type III secretion protein, induces toll-like receptor-independent apoptosis in macrophage through NFkappaB inhibition. J Biol Chem 2006; 281:36897 - 36904
- Burdette DL, Yarbrough ML, Orvedahl A, Gilpin CJ, Orth K. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proc Natl Acad Sci USA 2008; 105:12497 - 12502
- Kodama T, Rokuda M, Park KS, Cantarelli VV, Matsuda S, Iida T, et al. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol 2007; 9:2598 - 2609
- Bhattacharjee RN, Park KS, Chen X, Iida T, Honda T, Takeuchi O, et al. Translocation of VP1686 upregulates RhoB and accelerates phagocytic activity of macrophage through actin remodeling. J Microbiol Biotechnol 2008; 18:171 - 175
- Trosky JE, Mukherjee S, Burdette DL, Roberts M, McCarter L, Siegel RM, et al. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J Biol Chem 2004; 279:51953 - 51957
- Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci USA 2007; 104:17117 - 17122
- Yarbrough ML, Li Y, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 2009; 323:269 - 272
- Casselli T, Lynch T, Southward CM, Jones BW, DeVinney R. Vibrio parahaemolyticus inhibition of Rho family GTPase activation requires a functional chromosome I type III secretion system. Infect Immun 2008; 76:2202 - 2211
- Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol 2007; 10:24 - 29
- Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 2006; 62:631 - 640
- Bajaj V, Hwang C, Lee CA. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol 1995; 18:715 - 727
- Eichelberg K, Galan JE. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and hilA. Infect Immun 1999; 67:4099 - 4105
- Walker KA, Miller VL. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J Bacteriol 2004; 186:4056 - 4066
- Hovey AK, Frank DW. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol 1995; 177:4427 - 4436
- McCaw ML, Lykken GL, Singh PK, Yahr TL. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol 2002; 46:1123 - 1133
- Thibault J, Faudry E, Ebel C, Attree I, Elsen S. The anti-activator ExsD forms a 1:1 complex with ExsA to inhibit transcription of type III secretion operons. J Biol Chem 2009;
- Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 2004; 53:297 - 308
- Zhou X, Shah DH, Konkel ME, Call DR. Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol Microbiol 2008; 69:747 - 764
- Brutinel ED, Vakulskas CA, Yahr TL. ExsD inhibits expression of the Pseudomonas aeruginosa type III secretion system by disrupting ExsA self-association and DNA binding activity. J Bacteriol 2009; 192:1479 - 1486
- Lykken GL, Chen G, Brutinel ED, Chen L, Yahr TL. Characterization of ExsC and ExsD self-association and heterocomplex formation. J Bacteriol 2006; 188:6832 - 6840
- Yahr TL, Frank DW. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol 1994; 176:3832 - 3838
- Yahr TL, Hovey AK, Kulich SM, Frank DW. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol 1995; 177:1169 - 1178
- Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol 2004; 4:44
- Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescencebased reporter. J Bacteriol 2004; 186:5486 - 5495
- Croxatto A, Lauritz J, Chen C, Milton DL. Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ Microbiol 2007; 9:370 - 382
- Hakes DJ, Dixon JE. New vectors for high level expression of recombinant proteins in bacteria. Anal Biochem 1992; 202:293 - 298
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 1993; 130:127 - 130
- Milton DL, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J Bacteriol 1992; 174:7235 - 7244
- Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol 1996; 178:1310 - 1319
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 1991; 97:39 - 47