Abstract
Cryptococcus neoformans is unusual among melanotic fungi in that it requires an exogenous supply of precursor to synthesize melanin. C. neoformans melanizes during mammalian infection in a process that presumably uses host-supplied compounds such as catecholamines. L-3,4-dihydroxyphenylalanine (L-DOPA) is a natural catecholamine that is frequently used to induce melanization in C. neoformans and L-DOPA-melanized cryptococci manifest resistance to radiation, phagocytosis, detergents and heavy metals. Given that C. neoformans needs exogenous substrate for melanization one question in the field is the extent to which melanin-associated phenotypes reflect the presence of melanin or metabolic changes in response to substrates. In this study we analyze the response of C. neoformans to L-DOPA with respect to melanization, gene expression and metabolic incorporation. Increasing the concentration of L-DOPA promotes melanin formation up to concentrations > 1 mM, after which toxicity is apparent as manifested by reduced growth. The timing of C. neoformans cells to melanization is affected by growth phase and cell density. Remarkably, growth of C. neoformans in the presence of L-DOPA results in the induction of relatively few genes, most of which could be related to stress metabolism. We interpret these results to suggest that the biological effects associated with melanization after growth in L-DOPA are largely due to the presence of the pigment. This in turn provides strong support for the view that melanin contributes to virulence directly through its presence in the cell wall.
Introduction
Many pathogenic fungi produce melanin in the cell wall. Melanin enhances the virulence of fungi in various ways. In the rice blast fungus, Magnaporthe grisea, melanin strengthens the cell wall, allowing for the buildup of high turgor pressure that aids in invasion of plant tissues.Citation1 In Paracoccidioides brasiliensis, melanin interferes with normal macrophage function.Citation2,Citation3 In Exophiala dermatitidis, melanin contributes to fungal resistance to oxidative killing by neutrophils.Citation4 In addition melanin increases the resistance of fungi to environmental damage, as in the melanized fungi isolated from the radioactively contaminated Chernobyl reactors.Citation5 Melanin is one of the major virulence factors in Cryptococcus neoformans, a fungus that is a relatively frequent cause of life-threatening meningoencephalitis in immunocompromised individuals.Citation6 Cryptococcosis is one of the more common secondary infections in AIDS patients. There are estimated to be nearly 1 million cases of the disease each year, the majority of which are in sub-Saharan Africa.Citation7
In contrast to most other melanotic fungi that are able to make melanin from endogenous precursors, the synthesis of melanin by C. neoformans requires the addition of exogenous substrate in the medium, such as L-DOPA.Citation8 The initial chemical step in the synthesis of melanin is the oxidation of L-DOPA to dopaquinone and is catalyzed by laccase.Citation9 Subsequent steps are believed to occur spontaneously, producing dihydroxyindole or dihydroxyindole-2-carboxylic acid intermediates that finally polymerize into melanin.Citation10,Citation11 Melanins are amorphous polymers composed of crosslinked phenolic and/or indolic subunits.Citation12 Since the exact composition of subunits in the polymer has not been determined, the definition of a compound as a melanin is based on physical characteristics, including dark color, insolubility in most solvents, resistance to acid, oxidation by hydrogen peroxide and degradation by alkali.Citation13 In C. neoformans, melanin has a complex architecture with multiple layers and a granular surface.Citation14
Despite its importance to pathogenesis, many details of the cell biology of C. neoformans melanization remain unknown. In fact, the inability of C. neoformans to produce its own melanin without exogenous substrate makes this organism an ideal system to study melanization since it is possible to vary conditions to better understand this process. In this study we have explored the conditions that lead to optimum pigment expression and the response of the organism to the presence of substrate. The results provide new insights into the process of cryptococcal melanization that are relevant to understanding the role of melanin in pathogenesis.
Results
Dose-dependent responses of melanization and growth to L-DOPA.
A dose-response assay was carried out with L-DOPA to ascertain the effects on growth and melanization and determine the optimal concentration for the other experiments in the study (). Cells were plated in agar containing 0.1, 1.0 and 10 mM L-DOPA and examined visually for growth and melanization effects over several days. The highest concentration of L-DOPA inhibited growth of cells, producing smaller colonies. No growth differences were observed for cells grown in low or intermediate L-DOPA concentrations. Yeast grown on media with 1 mM L-DOPA produced the darkest colonies. At the highest concentration, less melanization occurred. A concentration of 1 mM L-DOPA was used in the subsequent experiments because it produced the most pigment without compromising growth of cells. Toxicity of the catecholamine norepinephrine was also tested. At a concentration of 10 mM, growth of C. neoformans was completely inhibited.
Timing of melanization.
When cells were cultured in liquid medium, we noted that it often took several days for the cells to turn black, depending on the strain. We reasoned that this delay in melanization could have several explanations. For example, the melanization lag could have indicated that L-DOPA was incorporated very slowly into melanin. Alternatively, it could have indicated that the melanization process took several days to commence, but occurred rapidly once begun. To distinguish between these possibilities, cells were grown for seven days in minimal medium without L-DOPA, and then L-DOPA was added to a concentration of 1 mM. After one or two days, the “old” cultures melanized (). In contrast, when cells were cultured for two days in “new” media containing L-DOPA, they did not melanize. This result suggested that the rate of melanization in culture was limited by the initiation of melanization, not the production of melanin.
Density of cells influenced melanization.
The results with old cultures suggested that cell density and/or age impacted the rate of melanization, since older, denser cultures melanized rapidly and new cultures did not. Cell density effects were tested by concentrating an overnight culture 10- and 100-fold. L-DOPA was added and the cultures were incubated overnight. Only the densest culture turned black; no color change was observed for the less dense cultures ().
Similar to growth in liquid, the timing at which melanization of C. neoformans cells grown in agar became visually apparent was variable. To test if the timing of melanization in agar was related to density, L-DOPA agar was inoculated with variable amounts of C. neoformans. When grown in agar, melanization occurred faster when more CFUs were plated per culture dish ().
Genes induced by L-DOPA.
Gene expression in melanizing cells was analyzed. L-DOPA was added to a culture of C. neoformans grown to a cell density of 4 × 107 CFU/ml. After four hours of incubation with L-DOPA, RNA was extracted from the cells and purified. Microarray analysis was carried out comparing cells grown in the presence of L-DOPA to those without L-DOPA. Of the approximately 6,500 genes in C. neoformans, eight were identified with increased expression in the presence of L-DOPA (). Real-time PCR was used to confirm the induction of the genes. Induction of five of the eight genes was confirmed by real-time PCR analysis (CNB04110, CNK02300, CND03820, CNG04630 and CNB04240). To determine the putative functions of the induced genes, homology searches were carried out by entering the predicted protein sequence into the NCBI BLAST search webpage (www.ncbi.nlm.nih.gov/BLAST). Among these genes, several were putative enzymes with redox functions ().
To test the relationship of these genes to melanization, we determined their expression in the presence of L-DOPA under non-melanizing conditions. To do so, gene induction was measured by real-time PCR in a C. neoformans strain without laccase (LAC1 deletion strain 2E-TU and the complemented strain, 2E-TUC, of a related genetic background). The same five genes with confirmed increase in gene expression had little or no induction in the laccase deletion strain. The complemented strain had variable expression, perhaps due to differences in laccase expression between this strain and the wild-type ().
Next, cultures were incubated with L-DOPA for several days and changes in gene expression were evaluated over time (). The maximum levels of induction were considerably higher at these longer times in comparison to the previous experiments in which cultures were incubated with L-DOPA for 4 h (). For example, gene CNB04110 had a maximum induction of 350-fold after three days of incubation with L-DOPA. A similar pattern of induction was observed for most of the genes, with low expression on day 1, a peak of gene expression on day 3, and gradual decrease over time. This peak of gene expression coincided with the cultures reaching a density at which melanization occurs. Gene induction was generally not observed in the laccase mutant strain.
Uptake of L-DOPA by C. neoformans cells.
The gene expression studies using the LAC1 deletion strain suggested that melanization was required for induction of the genes in the presence of L-DOPA. To further address the issue of whether the gene expression changes were related to melanization, the ability of cells to incorporate L-DOPA was tested. Cells were incubated with 14C-labeled L-DOPA and the level of incorporation into the cells was determined by liquid scintillation counting. Initial experiments showed a low level of incorporation for all cells (data not shown). To increase the level of L-DOPA incorporation, cells were incubated in starvation media prior to adding 14C-L-DOPA, a condition known to induce melanization.Citation15 Incorporation was measured at 5 min, 1 h, 4 h and 23 h of incubation. After overnight incubation, the wild-type and laccase complemented strains accumulated much of the 14C-L-DOPA (77% and 70%, respectively), while the laccase deletion strain and heat-killed cells incorporated a minimal amount (8% and 5%, respectively) (). At all time points except for 5 min, cells of the JEC21 and laccase-complemented strains had significantly greater radiation incorporation than both the laccase deletion and heat killed cells (p < 0.001). These results indicate that only melanizing cells accumulated L-DOPA.
Discussion
Melanization is critical to the virulence of C. neoformans and other pathogenic fungi. However, the cellular events that occur in melanizing cells are not well understood. The purpose of this study was to gain insight into the metabolism of L-DOPA into melanin by examining the response of C. neoformans to incubation with L-DOPA with regard to the timing of melanization and the changes in gene regulation.
Melanization of C. neoformans is affected by cell density. When grown in liquid medium with L-DOPA, C. neoformans cells require several days to a week to turn black. However, when cells are grown for several days prior to the addition of L-DOPA, melanization occurs rapidly after substrate addition. In general, the appearance of dark color in liquid culture coincides with cell densities of approximately 4 × 107 CFU/ml. Cell density effects are similarly apparent when cells are plated on solid medium. Melanization occurs more rapidly as the number of colonies per unit area increases. Together, these results suggest that the rate of melanization is density-dependent. This dependence of melanization on cell density has echoes of a quorum sensing effect. Indeed, these results add to considerable work that has been done in our labCitation16,Citation17 suggesting that C. neoformans melanization is controlled by quorum sensing. In this regard, a quorum sensing-like phenomenon has been described for C. neoformans mediated by a peptide.Citation18 Although this peptide has not been studied for its effect on the rate of melanization, it has protean changes in C. neoformans metabolism that include effects on melanizationCitation19 and it is conceivable that a related mechanism is responsible for our results reported in this manuscript.
Both melanization and growth are inhibited in the presence of a high concentration of L-DOPA or norepinephrine (10 mM). The inhibition of melanization may be due to autopolymerization of the substrate in the medium making it unavailable for use in melanin formation. Examination of the plates shows that the agar surrounding the colonies is dark at this concentration. The inhibition of growth suggests that L-DOPA, or a metabolic product of L-DOPA, presents some toxicity to C. neoformans. This hypothesis is supported by the gene expression data, as discussed below.
These data elucidate the optimal conditions for in vitro melanization of C. neoformans. The relationship between in vitro and in vivo melanization is an important topic about which very little is known. Melanized cryptococcal cells have been observed in the brains of patients as well as in experimental animals.Citation20,Citation21 The in vivo substrate has yet to be identified, but it is hypothesized to be one or a combination of catecholamines such as dopamine, norepinephrine, homovanillic acid and L-DOPA. Several of these compounds are normally present at concentrations of µg/g in brain tissueCitation22,Citation23 and nM in the CNS.Citation24 The distribution of catecholamines in the brain is not uniform. For example, dopamine concentration is particularly high in the basal ganglia,Citation25 a region where C. neoformans is often found.Citation26 The levels of these catecholamines in cryptococcosis patients are not known. Likewise, cell density effects on in vivo melanization are hard to determine. In vivo, cryptococcal cells may not be evenly distributed as in a liquid culture, but may be inside host macrophagesCitation27 or a cryptococcoma.Citation28 The effects of immune attack and hypoxia on C. neoformans further complicate the scenario.
One of the unanswered questions in the field is the extent to which growth of C. neoformans in media containing substrate affects fungal metabolism independently of the capacity of substrate to induce melanization. Published studies in melanization have identified genes required for C. neoformans to produce pigment, resulting in a white phenotype when mutated. These include genes in cell wall synthesis, metal ion transport and virulence regulators.Citation29,Citation30 In contrast, our study identifies novel genes that are upregulated in the presence of melanization substrate. Gene induction is only observed in cells capable of melanization, suggesting the genes either have a role in this process or are induced as a consequence of melanization. In addition, the density at which maximal gene expression occurs is similar to the density at which melanization becomes visible in liquid culture. Furthermore, analysis of L-DOPA incorporation by C. neoformans cells shows that only wild-type cells capable of melanization incorporate the substrate. This suggests that L-DOPA does not accumulate inside cells and is unlikely to have an effect on gene expression that is unrelated to melanization. Together, these data support the hypothesis that the identified genes play a role in melanization.
We hypothesize that the genes identified function in regulating melanization and/or protecting cells from toxic by-products produced during melanization. In general, melanin and its synthetic intermediates are highly reactive molecules. For example, the intermediate dopaquinone can react with amine and thiol groups, such as in proteins, resulting in crosslinking of melanin and other molecules.Citation31 Thus, it seems logical that melanization is regulated in some way. The toxicity hypothesis is supported by published data showing that two of the genes, CNB04110 and CNB04240, are also induced by nitric oxide stress.Citation32 Furthermore, homology searches also provided information consistent with the toxicity hypothesis. CNG04630 has homology to permeases of the major facilitator family, proteins involved in removal of toxic substances from the cell, whereas CNK02300 has homology to glutathione transferases, proteins involved in metabolizing toxins inside cells.
Perhaps the most surprising aspect of the gene expression analysis is the paucity of genes that are differentially regulated in the melanized state. Given that melanization is associated with diverse new attributes for melanized cells such as resistance to radiation,Citation33 heavy metals,Citation34 oxidants,Citation35,Citation36 enzymatic degradation,Citation37 antifungal drugsCitation38,Citation39 and defensins,Citation40 one might have expected melanin synthesis to be associated with more global changes in gene expression. In fact, the relative paucity of gene expression changes implies that melanization in C. neoformans is a relatively simple system from a genetic point of view and this, in turn, supports the association of new cellular properties in melanized cells with the formation of pigment rather than substrate-induced metabolic changes. The paucity of gene responses is also consistent with our recent proposal that melanin is synthesized in vesicles that are subsequently exported to the cell wallCitation41 since this model involves pigment synthesis from highly reactive and toxic intermediates of L-DOPA oxidation in a confined membrane enclosed compartment that shields cellular processes. A similar strategy for avoiding melanin precursor toxicity is used by mammalian cells, which make melanins within melanosomes. In this regard, defective melanosomes have been associated with necrosis in melanoma cells.Citation42 Nevertheless, the observation that most differentially regulated genes are associated with redox functions suggests that C. neoformans growth in the presence of L-DOPA is a stressful condition for this microbe. In summary, our results show that melanization in C. neoformans is a cell density dependent process that is associated with the induction of very few genes relative to the protean effects conferred by pigmentation that reduce fungal cell susceptibility to numerous insults. This in turn provides strong evidence for the view that the pigment itself is responsible for many protective effects associated with melanization and further strengthens the connection between melanin synthesis and virulence.
Materials and Methods
Fungal strains.
JEC21 is a serotype D, MATα strain that was produced by crossing an environmental isolate with a clinical isolate.Citation43 The serotype A, MATα strain H99 is a clinical isolate.Citation44 The serotype D LAC1 deletion strain (2E-TU) contains a partial deletion of LAC1 near the 5′ end of the gene, and the complemented strain (2E-TUC) contains an integrated copy of LAC1.Citation45 The laccase deletion and complemented strains are related to JEC21.
Growth and melanization assays.
C. neoformans cells were grown in chemically defined minimal medium (15 mM dextrose, 10 mM MgSO4, 29.4 mM KH2PO4, 13 mM glycine and 3 µM thiamine, pH 5.5) with or without 1 mM L-DOPA (Sigma-Aldrich) and incubated at 30°C, 150 rotations per minute (RPM) in the dark. Cultures were examined daily to monitor growth and pigment production.
Cell density assays.
Cell densities were determined by hemocytometer count and/or plating on agar. For the plate assay, the indicated numbers of CFUs were plated on chemically defined minimal medium agar containing 1 mM L-DOPA. For the liquid assay, an overnight culture grown in chemically defined minimal medium at a density of 3 × 105 CFU/ml was concentrated 10- and 100-fold. L-DOPA (1 mM) was added to the concentrated cultures and they were incubated for 18 h before photographing. All incubations were done at 30°C, 150 RPM in the dark.
Microarray analysis of genes induced by L-DOPA.
C. neoformans cells were grown in minimal medium without substrate at 30°C, 150 RPM to a density of about 4 × 107 CFU/ml. L-DOPA was then added at a concentration of 1 mM and the cells were allowed to grow for an additional four hours. After the incubation, cells grown in the presence of L-DOPA were slightly grayish in color, indicating that melanization was occurring in the cells. Two independent biological replicates were performed. Total RNA was isolated from the cultures using the RNeasy kit (Qiagen) and incubated with DNase (GenHunter) to remove contaminating DNA. The purified RNA was sent to Washington University Genome Sequencing Center for microarray analysis with the JEC21 genomic microarray as follows. Comparisons were done with both RNA pools and a Cy3–Cy5 dye swap was done between the sample with and without L-DOPA. Immediately after hybridization, the microarray slides were scanned on a ScanArray Express HT Scanner (Perkin Elmer) to detect fluorescence. Gridding and analysis of images was performed with ScanArray Software Express V2.0 (Perkin Elmer). Gene expression data were averaged across the two RNA pools and analyzed using the GeneSpring 7.2 software (Agilent) to identify genes in which the mean of the replicates had >2-fold change and p < 0.05. The Benjamini & Hochberg false discovery rate for multiple testing corrections was then done for all genes showing changes in gene expression.Citation46
Real-time PCR analysis of identified genes.
L-DOPA was added to C. neoformans cells of strain JEC21, 2E-TU or 2E-TUC at densities of between 2 and 5 × 107 CFU/ml and RNA was isolated after four hours as described above. cDNA was made by reverse transcription of two independent pools of RNA using the Quantitect Reverse Transcription kit (Qiagen). For the time course analysis, L-DOPA was added to cells at a density of 1 × 104 CFU/ml (day 0) and RNA was isolated days 1, 3, 5 and 8. Primers were designed based on the JEC21 genome sequences to amplify approximately 100 base pairs (). PCR products were amplified with SYBR® Green PCR Master Mix (Applied Biosystems) in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Fold change in gene expression was determined relative to untreated cells of the same strain according the method of Pfaffl.Citation47 The gene encoding glyceraldehyde-3-phosphate dehydrogenase was used as the reference.
[14C]L-DOPA incorporation analysis.
Cells of C. neoformans strains JEC21, 2E-TU and 2E-TUC were cultured for two days in chemically defined minimal medium at 30°C, 150 RPM. Cells were collected by centrifugation, washed and suspended in starvation medium (0.2 g/l, K2HPO4, 0.1 g/l KH2PO4). The cells were then incubated overnight at 30°C and 150 RPM. Before incubation with the labeled substrate, cells were collected by centrifugation again and suspended in starvation medium to a final density of 1–2 × 108 CFU/ml. Heat-killed cells were incubated at 65°C for 1 h. Cell suspensions were incubated with L-3,4-dihydroxyphenyl[3-14C]alanine [4 µCi of a 54 mCi/mmol solution (GE Healthcare)] such that the concentration of L-DOPA was approximately 0.005 mM. At indicated time points, aliquots were removed and briefly centrifuged to pellet cells. The pellets were washed twice with PBS to remove unincorporated label. Liquid scintillation counting was used to determine the total counts per minute (CPM) in the supernatants and pellets (LKB Wallac 1217 Rackbeta, PerkinElmer). Percent incorporation was calculated using the formula: CPM pellet ÷ (CPM pellet + CPM supernatant) × 100. A multivariate analysis of variance (MANOVA) was used to test for incorporation differences between C. neoformans strain and time. Simple contrasts were then done to determine significant differences among strains at a particular time.
Figures and Tables
Figure 1 Melanization dose response of C. neoformans to L-DOPA and Norepinephrine. Cells of strains JEC 21 (top row) and H99 (bottom row) were serially diluted and plated on media with or without the indicated concentration of substrate (mM). Plates were incubated for four (L-DOPA) or seven (Norepinephrine) days before photographing.
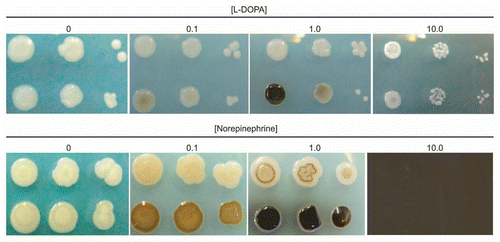
Figure 2 Effect of culture age on the rate of L-DOPA-mediated melanization. “New” indicates L-DOPA was added to cultures at the time of inoculation and incubated two days. “Old” indicates L-DOPA was added to a seven-day culture of C. neoformans and incubated for one or two more days. Left tube, strain JEC 21; right tube, strain H99.
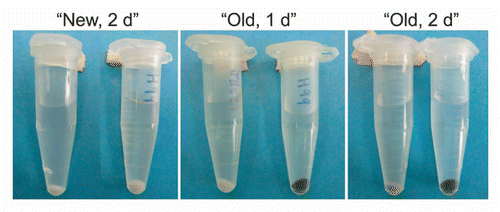
Figure 3 Effect of cell density on melanization. (A) C. neoformans strain JEC 21 was grown overnight in chemically defined minimal medium and concentrated before adding L-DOPA. Flasks were photographed after overnight incubation with L-DOPA. Cell densities of the cultures before adding L-DOPA were 6.6 × 105, 4.6 × 106 and 3.6 × 107 CFU/ml. (B) L-DOPA agar was inoculated with 12, 135 or 1,250 CFUs of C. neoformans strain JEC 21 (top) or 14, 90 and 1,400 CFU of strain H99 (bottom) and incubated seven days at 30°C.
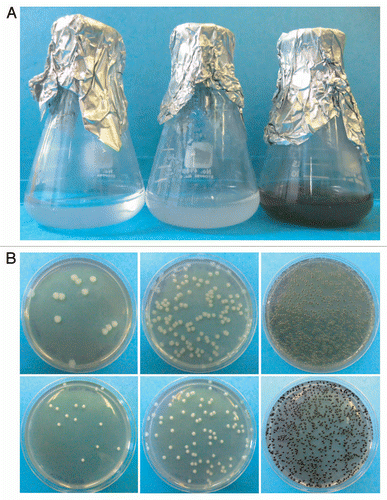
Figure 4 Fold changes in gene expression over time. C. neoformans wild-type (top) and laccase deletion (bottom) cultures were incubated with or without L-DOPA for 1–8 days. RNA was isolated from the cells at the indicated times for gene expression analysis by real-time PC R. The experiment was repeated twice. The results of one representative time course are shown. The fold changes in gene expression in the presence of L-DOPA are indicated for each gene.
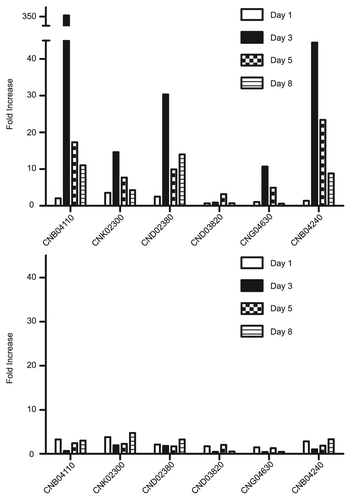
Figure 5 Incorporation of L-DOPA by cells. Assays were performed as described in the Methods and the percentage of L-DOPA incorporated calculated at indicated time intervals. Strains used were JEC 21 (WT), 2E-TU (Δlac1), 2E-TUC (COMPL.) and heated killed JEC 21 (HK).
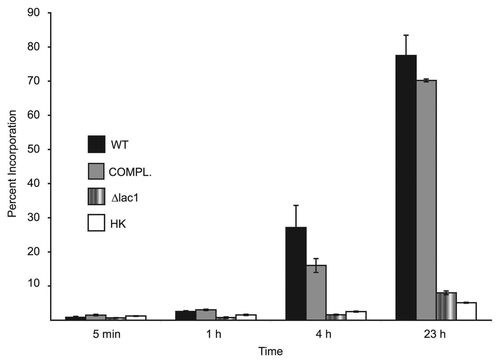
Table 1 Primers used for real-time PCR amplification
Table 2 Fold changes in gene expression upon addition of L-DOPA to cultures
Acknowledgments
This work was supported (in part) by a grant from The City University of New York PSC-CUNY Research Award Program to H. Eisenman. A. Casadevall is supported by NIH grants HL059842, AI033774, AI033142 and AI052733. In addition, we thank the Washington University Genome Sequencing Center for performing the microarray analysis.
References
- Howard RJ, Valent B. Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu Rev Microbiol 1996; 50:491 - 512
- Bocca AL, Brito PP, Figueiredo F, Tosta CE. Inhibition of nitric oxide production by macrophages in chromoblastomycosis: a role for Fonsecaea pedrosoi melanin. Mycopathologia 2006; 161:195 - 203
- da Silva MB, Marques AF, Nosanchuk JD, Casadevall A, Travassos LR, Taborda CP. Melanin in the dimorphic fungal pathogen Paracoccidioides brasiliensis: effects on phagocytosis, intracellular resistance and drug susceptibility. Microbes Infect 2006; 8:197 - 205
- Schnitzler N, Peltroche-Llacsahuanga H, Bestier N, Zundorf J, Lutticken R, Haase G. Effect of melanin and carotenoids of Exophiala (Wangiella) dermatitidis on phagocytosis, oxidative burst and killing by human neutrophils. Infect Immun 1999; 67:94 - 101
- Zhdanova NN, Zakharchenko VA, Vember VA, Nakonechnaya LT. Fungi from Chernobyl: mycobiota of the inner regions of the containment structures of the damaged nuclear reactor. Mycol Res 2000; 104:1421 - 1426
- Casadevall A, Steenbergen JN, Nosanchuk JD. ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi—the Cryptococcus neoformans paradigm. Curr Opin Microbiol 2003; 6:332 - 337
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 2009; 23:525 - 530
- Kwon-Chung KJ, Tom WK, Costa JL. Utilization of indole compounds by Cryptococcus neoformans to produce a melanin-like pigment. J Clin Microbiol 1983; 18:1419 - 1421
- Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol 1994; 176:656 - 664
- Land EJ, Riley PA. Spontaneous redox reactions of dopaquinone and the balance between the eumelanic and phaeomelanic pathways. Pigment Cell Res 2000; 13:273 - 277
- Williamson PR. Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front Biosci 1997; 2:99 - 107
- Wakamatsu K, Ito S. Advanced chemical methods in melanin determination. Pigment Cell Res 2002; 15:174 - 183
- Nosanchuk JD, Casadevall A. The contribution of melanin to microbial pathogenesis. Cell Microbiol 2003; 5:203 - 223
- Eisenman HC, Nosanchuk JD, Webber JB, Emerson RJ, Camesano TA, Casadevall A. Microstructure of cell wall-associated melanin in the human pathogenic fungus Cryptococcus neoformans. Biochemistry 2005; 44:3683 - 3693
- Chaskes S, Edberg SC, Singer JM. A DL-DOPA drop test for the identification of Cryptococcus neoformans. Mycopathologia 1981; 74:143 - 148
- Albuquerque P. 2011; Bronx, NY Albert Einstein College of Medicine Ph.D. Thesis
- Albuquerque P, Nicola A, Williamson PR, Casadevall A. Quorum sensing regulates growth and virulence in Cryptococcus neoformans 2011; 8th International Conference on Cryptococcus and Cryptococcosis Charleston, South Carolina
- Lee H, Chang YC, Nardone G, Kwon-Chung KJ. TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Mol Microbiol 2007; 64:591 - 601
- Lee H, Chang YC, Varma A, Kwon-Chung KJ. Regulatory diversity of TUP1 in Cryptococcus neoformans. Eukaryot Cell 2009; 8:1901 - 1908
- Nosanchuk JD, Rosas AL, Lee SC, Casadevall A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 2000; 355:2049 - 2050
- Rosas AL, Nosanchuk JD, Feldmesser M, Cox GM, McDade HC, Casadevall A. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect Immun 2000; 68:2845 - 2853
- Carlsson A. The occurrence, distribution and physiological role of catecholamines in the nervous system. Pharmacol Rev 1959; 11:490 - 493
- Glowinski J, Iversen LL. Regional studies of catecholamines in the rat brain. I. The disposition of [3H]norepinephrine, [3H]dopamine and [3H]dopa in various regions of the brain. J Neurochem 1966; 13:655 - 669
- Tohgi H, Abe T, Saheki M, Yamazaki K, Murata T. Concentration of catecholamines and indoleamines in the cerebrospinal fluid of patients with vascular parkinsonism compared to Parkinson's disease patients. J Neural Transm 1997; 104:441 - 449
- Adolfsson R, Gottfries CG, Roos BE, Winblad B. Post-mortem distribution of dopamine and homovanillic acid in human brain, variations related to age, and a review of the literature. J Neural Transm 1979; 45:81 - 105
- Klock C, Cerski M, Goldani LZ. Histopathological aspects of neurocryptococcosis in HIV-infected patients: autopsy report of 45 patients. Int J Surg Pathol 2009; 17:444 - 448
- Feldmesser M, Tucker S, Casadevall A. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol 2001; 9:273 - 278
- Li Q, You C, Liu Q, Liu Y. Central nervous system cryptococcoma in immunocompetent patients: a short review illustrated by a new case. Acta Neurochir (Wien) 152:129 - 136
- Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol 2005; 57:1381 - 1396
- Zhu X, Williamson PR. A CLC-type chloride channel gene is required for laccase activity and virulence in Cryptococcus neoformans. Mol Microbiol 2003; 50:1271 - 1281
- Land EJ, Ramsden CA, Riley PA. Quinone chemistry and melanogenesis. Methods Enzymol 2004; 378:88 - 1109
- Missall TA, Pusateri ME, Donlin MJ, Chambers KT, Corbett JA, Lodge JK. Posttranslational, translational and transcriptional responses to nitric oxide stress in Cryptococcus neoformans: implications for virulence. Eukaryot Cell 2006; 5:518 - 529
- Wang Y, Casadevall A. Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Appl Environ Microbiol 1994; 60:3864 - 3866
- Garcia-Rivera J, Casadevall A. Melanization of Cryptococcus neoformans reduces its susceptibility to the antimicrobial effects of silver nitrate. Med Mycol 2001; 39:353 - 357
- Jacobson ES, Tinnell SB. Antioxidant function of fungal melanin. J Bacteriol 1993; 175:7102 - 7104
- Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun 1994; 62:3004 - 3007
- Rosas AL, Casadevall A. Melanization decreases the susceptibility of Cryptococcus neoformans to enzymatic degradation. Mycopathologia 2001; 151:53 - 56
- Ikeda R, Sugita T, Jacobson ES, Shinoda T. Effects of melanin upon susceptibility of Cryptococcus to antifungals. Microbiol Immunol 2003; 47:271 - 277
- van Duin D, Casadevall A, Nosanchuk JD. Melanization of Cryptococcus neoformans and Histoplasma capsulatum reduces their susceptibilities to amphotericin B and caspofungin. Antimicrob Agents Chemother 2002; 46:3394 - 3400
- Doering TL, Nosanchuk JD, Roberts WK, Casadevall A. Melanin as a potential cryptococcal defence against microbicidal proteins. Med Mycol 1999; 37:175 - 181
- Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology 2009; 155:3860 - 3867
- Borovansky J, Mirejovsky P, Riley PA. Possible relationship between abnormal melanosome structure and cytotoxic phenomena in malignant melanoma. Neoplasma 1991; 38:393 - 400
- Heitman J, Allen B, Alspaugh JA, Kwon-Chung KJ. On the origins of congenic MATalpha and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet Biol 1999; 28:1 - 5
- Perfect JR, Ketabchi N, Cox GM, Ingram CW, Beiser CL. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol 1993; 31:3305 - 3309
- Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med 1996; 184:377 - 386
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 1995; 57:289 - 300
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:45