Abstract
Biofilms are masses of microorganisms that bind to and multiply on a solid surface, typically with a fluid bathing the microbes. The microorganisms that are not attached but are free floating in an aqueous environment are termed planktonic cells. Traditionally, microbiology research has addressed results from planktonic bacterial cells. However, many recent studies have indicated that biofilms are the preferred form of growth of most microbes and particularly those of a pathogenic nature. Biofilms on animal hosts have significantly increased resistance to various antimicrobials compared to planktonic cells. These microbial communities form microcolonies that interact with each other using very sophisticated communication methods (i.e., quorum-sensing). The development of unique microbiological tools to detect and assess the various biofilms around us is a tremendously important focus of research in many laboratories. In the present review, we discuss the major biofilm mechanisms and the interactions among oral bacteria.
Introduction
Anton van Leeuwenhoek was the first scientist who observed microorganisms with a microscope in 1683.Citation1 One of the first samples he examined was his own dental plaque or biofilm. Ever since then, microorganisms began to attract researchers' interests. In the beginning, scientists mainly focused on single bacterial species performance in broth culture. This type of bacterial growth is termed planktonic growth. But later, scientists realized that most, if not all, bacteria live in biofilms instead of broth. Therefore, biofilm investigations drew scientists' attention, and this area of research has become popular in recent years. Biofilms are defined as orientated aggregations of microorganisms attached to each other or to a surface and enclosed in extracellular polymeric substance (EPS) produced by themselves.Citation2–Citation5 Furthermore, many biofilms are bathed by some flowing fluid (i.e., water flowing over moss-covered rocks in a stream or saliva flowing over dental plaque on a tooth surface). The components of mature biofilm are approximately 5–25% bacterial cells and 75–95% glycocalyx matrix.Citation6
Biofilms can be found almost everywhere associated with moisture. It is well known that there are at least 10 times more bacteria inhabiting the human body than the number of human cells. In nature, biofilms are found in industrial bioreactors, on rocks in streams, and in animal host environments such as in or on the otolaryngologic, vaginal and gastrointestinal tracts. Otolaryngologic biofilms are typically constituted by Pseudomonas aeruginosa-containing EPS-cellular towers that are separated by open passages that deliver nutrients and dispose of metabolic wastes. The classical mushroom-type biofilm structures formed by P. aeruginosa contain one pseudomonal strain in the stalks and a second different strain in the mushroom heads.Citation7 The human gastrointestinal tract contains a rich and diverse microbiota along its length and the study of the gastrointestinal tract microbiome is a rapidly growing field. A large number of intestinal bacterial species on the intestinal mucosa surface form dense biofilms. There are a total of about 1014 intestinal bacterial species, mainly in the colon.Citation8 In the gastrointestinal tract, most microorganisms colonize the large intestine. Gut biofilm is composed of both living and dead bacteria in the mucus layer lining mucosal surfaces or on food residues in the lumen of the gut. Bacteroides and bifidobacteria are the predominant bacterial species attaching to particulate matter in stool and were shown to be phenotypically similar to the nonadherent microbiota. Vaginal bacterial groups in healthy women are generally divided into resident bacteria, non-resident bacteria and occasionally resident bacteria. Microorganisms in the healthy vagina maintain balance with the host and the environment. This process is called ecological balance. Lactobacilli are dominant in the vaginal microorganisms of a healthy woman, although other bacteria can be present in lower numbers.Citation9 Therefore, a reduction in the numbers of vaginal lactobacilli can lead to vaginal flora imbalance, decrease in vaginal cleanliness and abnormal pH, so that harmful microorganisms are increased and the large population of endogenous bacteria are decreased.
There are over 700 different bacterial species in the oral microflora.Citation2–Citation5 Those species colonize the teeth, tongue, oral mucosa, hard palate, carious lesions, periodontal pocket, et al. The distribution of microflora in the oral cavity is not random, most species prefer certain sites over others due to the particular local environment those sites provide, such as the anaerobic environment provided by the periodontal pocket.Citation10 The majority of the microflora benefit our health while the minority are harmful, however, most research focuses only on those pathogenic species giving the public an illusion that bacteria are our “enemies.” Actually, most bacteria are “friendly” commensals and even essential components to our health. Roberts and DarveauCitation11 proposed that dental biofilm and their products contribute to healthy periodontium. Microflora can be found in both caries-free and periodontitis-free people and caries-affected and periodontitis-affected people, and many clinical studies reveal that the portion of certain bacterial species such as Streptococcus mutans or Porphyromonas gingivalis, respectively, is increased in patients with caries or periodontitis. Therefore, it seems that the competition that results between beneficial bacteria and virulent bacteria leads to either a healthy or sick status of human beings. Competition between members of the dental microflora are very complex and many antagonistic characteristics can be observed from competition for initial attachment on tooth surfaces or for later attachment to pioneer bacteria, competition from bacteriocins or hydrogen peroxide secreted and from facilitating the growth of some species which inhibit other species. To date only some of the details of these mechanisms are known. The present review will provide an overview of the formation of dental biofilm, the major mechanisms of oral bacterial interactions and inter-species bacterial interactions. Due to the large number of oral bacterial species, only the best characterized species are included in this review.
Oral Biofilm Formation
Acquired pellicle formation.
Oral biofilm is unique among the various types of biofilms as it typically requires host salivary glycoproteins to attach to. The first step of oral biofilm formation is the attachment of acquired pellicle, which is a thin protein-containing film derived from salivary glycoproteins, to a clean tooth surface (). The mechanism of acquired pellicle formation is based on Gibbs law of free enthalpy in that more energy will be released if glycoproteins are attached to the tooth surface. Many interactions among the various glycoproteins, other salivary components, and the tooth surface are involved. The forces of those interactions can be roughly divided into three types: long-range forces (50–100 nm between the two interacting components), medium-range forces (10–50 nm) and short-range forces (less than 5 nm). The long range forces include Coulomb interactions, van der Waals forces and dipole-dipole interactions; the medium range forces include hydrophobic interactions; and the short range forces include covalent bonds, electrostatic interactions, hydrogen bonds, ionic interactions and Lewis acid-base interactions.Citation12 With these forces, proteins are absorbed and rearranged, some conformational changes are undertaken,Citation13 and the new pellicle forms are ready for pioneer bacteria adhesion.
Initial adhesion.
Bacteria adhesion to the pellicle is the second step of biofilm formation. Some of the planktonic bacteria may recognize binding proteins in acquired pellicle, i.e., α-amylase and proline-rich glycoproteins/proteins and bind to the pellicle (). However, in this stage the attachment is reversible and those initially attached bacteria can detach from the pellicle easily. The early attachments are primarily based on electrostatic attractions or physical attachments,Citation14 but later, chemical forces become predominant. As soon as the pioneer bacteria attach to the pellicle, they begin to excrete EPS, which helps the bacteria stay bound together and attach to the pellicle. The main surface attachment structures of bacteria are fimbriae and fibrils. Fimbriae are composed of fimbrillins; while fibrils are like fimbriae, but they are shorter and thinner than fimbriae with more or less density distributed on the cell surface.Citation15 The forces present in the pioneer bacterial adhesion to the salivary acquired pellicle are hydrogen bonds, hydrophobic interactions, calcium bridges, van der Waals forces, acid-base interactions and electrostatic interactions.Citation12 Actinomyces spp, Streptococcus spp, Haemophilus spp, Capnocytophaga spp, Veillonella spp, and Neisseria are the main pioneer bacterial genera attaching to the tooth surface.Citation16–Citation18 Those early colonizations help those species occupy space and gain advantage in later competition with other species.Citation19
Maturation.
Since earlier reviews, such as Kolenbrander et al.,Citation4,Citation20 Hannig and Hannig,Citation12 and Hojo et al.Citation5 already provided very comprehensive and detailed summaries of biofilm maturation, in the present article we briefly discuss mature biofilm formation and coaggregations between bacteria. After the pioneer bacteria attach, those early colonizing bacteria provide specific binding sites either directly or through salivary glycoproteins binding to the pioneer organisms for subsequent bacterial colonization and promote the development of biofilm (). Later colonizing bacteria recognize polysaccharide or protein receptors on the pioneer bacterial cell surface and attach to them. Bacteria coaggregate, forming the typical corn cob forms, bristle brush forms, or other forms in mature oral biofilm. Subsequent attached bacterial species include Fusobacterium nucleatum, Treponema spp, Tannerella forsythensis, P. gingivalis, Aggregatibacter actinomycetemcomitans, et al.Citation17,Citation20 The microbial components of mature biofilm are quite different from the initial biofilm. A proportional shift occurs during biofilm development. The relative amount of Streptococci and Neisseria decrease, while the amount of Actinomyces, Corynebacterium, Fusobacterium and Veillonella increase.Citation16 Bacterial aggregation is a very complex process. Mature biofilms typically contain many porous layers and water channels through the biofilm, providing the bacteria essential nutrients.
Aggregation is the foundation of bacterial interactions in biofilm formation. Generally, one bacterial species can coaggregate with many other bacterial species by cell-to-cell recognition. Paired aggregation tests among 300 oral bacteria species indicated more than 90% undergo coaggregation.Citation21 However, aggregation is also specific because one bacterial strain cannot aggregate with any random bacterial strains, i.e., S. mutans aggregates with F. nucleatum but not with P. gingivalis. The fundamental mechanism of aggregation is polysaccharide recognition between bacteria. The polysaccharide recognition sites vary from one paired bacterial recognition to another paired bacterial recognition, because one bacterial cell has several different receptors which are complementary to different adhesions belonging to other bacterial species. Many aggregations are inhibited by lactose but not by sucrose. McIntireCitation22 reported the aggregation between Streptococcus and Actinomyces was 90% inhibited by lactose and D-galactose, but was only 10% inhibited by maltose, sucrose, cellobiose, or other monosaccharides. Coaggregation bridges usually refer to a structure of one bacterial species with two or more different receptors which can be recognized by different adhesions of two or more different bacterial species. Those bacterial species could aggregate by attaching to the first bacterial species; i.e., F. nucleatum is one of the best known coaggregation bridge species that facilitates streptococcal and obligate anaerobes aggregation. However, on the other hand, if two bacterial cells recognize the same receptor, the two cells would also compete for the binding site.
Dispersion of biofilm cells.
In mature biofilm, bacteria leave the biofilm by single cell detachment or a cluster of cells detaching (). The model of biofilm dispersion includes erosion, sloughing and seeding.Citation23 At least two possible reasons can be explained for the detachment. The first is due to limited nutrients present at the original site and bacteria have to locate a new site with more nutrients for growth. This nutrient-seeking reason could be further divided into active dispersion and passive dispersion. Active dispersion is conducted by the particular bacterium themselves, while passive dispersion is conducted by other bacterial species which compete for the nutrient with this particular bacteria. The second reason is host defense such as fluid shear force of saliva, which tries to limit biofilm development. For more detailed information about dispersion, please refer to Kaplan's review.Citation23
Cooperation in Biofilm
Almost all bacteria in nature grow in biofilm communities, where bacteria work as a well organized society. Bacterial cells in biofilm acquire distinguishing characteristics compared to planktonic cells. Previous studies in our labCitation24 demonstrated different protein expression in biofilm and planktonic S. mutans, in which fructan hydrolase, exo-beta-D-fructosidase, fructanase and cell division protein FtsG were enhanced in biofilm organisms compared to planktonic cells, but antigen I/II, glucosamine-fructose-6-phosphate aminotransferase I and chaperonin GroEL were repressed in biofilm cells. Some of our preliminary data indicate that in planktonic cells low concentrations of nicotine (0.25–0.5 mg/ml) enhanced S. mutans growth, while higher concentrations (2–4 mg/ml) of nicotine repressed S. mutans growth, but in biofilm both low and high concentrations of nicotine enhanced S. mutans growth (Huang, Li and Gregory, manuscript in preparation). For most bacterial species, biofilms are the preferred form of growth.Citation25 Biofilm provides many advantages to the reproduction, metabolism and defense of bacteria from other bacteria or the host.
Metabolic communication.
The foundation of biofilm metabolic communication is coaggregation, which provides cells close access and makes communication more convenient between bacterial species.
EPS, as mentioned above, is the major component of biofilms. All biofilms contain EPS, although the type of EPS in biofilms varies from one biofilm to the other due to the bacterial growth status and the substrates for bacterial metabolism. Because almost all bacterial species in biofilm can undergo biosynthesis and degrade EPS, EPS becomes the communication medium between bacterial species. Other functions of EPS are to provide shelter to the bacteria as the cross-linked EPS can block harmful agents outside of the biofilm and trap nutrients from the environment for a local higher concentration of the nutrients and extracellular enzymes for better biosynthesis of substrate. EPS helps biofilm maintain biofilm structures. Although the biofilm structures change according to the bacterial species in the biofilm and the nutrients available in the environment, EPS adapts to a new confirmation and becomes compatible with the requirement of the biofilm. EPS also influences iron exchange within biofilms and controls the hydrophilic or hydrophobic characteristics of the biofilm itself.
Oxygen metabolism and exchange within the biofilm between different aerobic and obligate anaerobic species has a special significant role for the survival of obligate anaerobes. It is difficult for obligate anaerobe species to live in an aerobic environment without the cooperation of aerobic species. The aerobic species consume oxygen in the environment, resulting in a lower level of oxygen concentration and producing a local redox potential gradation which provides local anaerobic conditions. The local anaerobic environment is the foundation of obligate anaerobic species survival. F. nucleatum is an important bridge bacterium, which can aggregate with both aerobic and obligate anaerobic species, allowing the two species to live together ().Citation26,Citation27 Without F. nucleatum, the number of obligate anaerobic species would decrease sharply in the oral cavity.Citation28
Some bacterial species can help other species survive by depleting acid and improving the local environment. F. nucleatum and Prevotella intermedia can grow in a wide pH range from 5.0 to 7.0, but P. gingivalis is very susceptible to a pH lower than 6.5. However, P. intermedia and F. nucleatum can produce ammonia and organic acids by fermenting glutamate and aspartate,Citation29 providing a more neutral environment for P. gingivalis and other acid-sensitive bacterial species.
Other metabolic communications between oral bacterial species, such as short-chain fatty acids, exogenous quinines and vitamin K, were thoughtfully reviewed by Hojo and colleagues.Citation5
Resistance to antibiotics.
Antibiotic resistance has become a worldwide problem in public health. The resistance of Streptococci to penicillin, amoxicillin, trimethoprimsulfamethoxazole and erythromycin was observed in children treated with antibiotics for otitis.Citation30 Biofilms can protect bacteria from a challenging environment with several host defense mechanisms directed towards bacteria or protect from applied antibiotics. The antigens of biofilm bacteria are hidden in the biofilm matrix and become less suspectible to the host immune system. Physical injury of the biofilm can also be reduced by the biofilm matrix. The short distance between bacterial cells makes cell-to-cell metabolic communications more frequent.Citation31 Scientists postulated a conception of “insurance hypothesis,” which stated a single species was more vulnerable by the environment than multiple species. If there are more species in the biofilm, and the structure of the biofilm is more complex and mature, the resistance to antibiotics is greater.Citation32 The antibiotic resistance of bacterial cells in biofilm was reported to be 1,000 to 1,500 times greater than the resistance of planktonic cellsCitation33 and has become a rising problem in recent years. Antibiotic resistance genes can be transferred between bacterial cells within biofilm resulting in a biofilm-wide resistance to the antibiotics.Citation34 The common carrier of resistance genes are plasmids, which are circular double-stranded DNA species presenting mainly in bacteria. The replication of plasmids is independent of chromosomal DNA replication and the number of plasmids in a cell varies widely. Plasmids with antibiotic resistance genes are gained by bacterial conjugation.
Sedlacek and WalkerCitation35 compared the antibiotic effect between biofilm and planktonic cells. There were 19 subgingival species investigated: Actinomyces naeslundii, Actinomyces meyeri, Bacteroides species, Bifidiobacterium species, Campylobacter species, Eubacterium species, Fusobacterium alocis, F. nucleatum, Faecalibacterium prausnitzii, P. intermedia/nigrescens, Prevotella loescheii, Streptococcus intermedius, Streptococcus parasanguis, Streptococcus sanguinis (earlier named Streptococcus sanguis), Veillonella atypical and Veillonella parvula. Both biofilm and planktonic cells were treated with tetracycline, amoxicillin and clindamycin with a concentration range from 0 to 2,048 µg/ml. Almost all biofilm cells displayed a higher minimum inhibitory concentration (MIC) compared to their relative planktonic cells. A. naeslundii, Campylobacter species, F. nucleatum, P. intermedia/nigrescens, P. loescheii, S. intermedius, S. parasanguis, S. sanguinis, V. atypical and V. parvula demonstrated very significant (4–250-fold) antibiotic resistance increases between biofilm and planktonic cells.
In addition, antibiotic resistance varies from one antibiotic to another antibiotic. Sedlacek and WalkerCitation35 found the concentrations to inhibit F. nucleatum growth are 256, 128 and 64 µg/ml for tetracycline, amoxicillin and clindamycin, respectively. Besides, antibiotic resistance varies from one bacterial species to another species. For example, the MIC of tetracycline on single species biofilm cells were 64 µg/ml for A. meyeri, but 512–2,048 µg/ml for A. naeslundii.Citation35
Competition in Biofilm
As a complex and diverse society, competition is highly prevalent among bacteria within biofilm. The bacterial species compete for nutrients, binding sites, and the chance to survive. Several major competitive mechanisms are wildly adopted by bacteria, i.e., bacteriocin synthesis, quorum sensing and hydrogen peroxide excretion.
Bacteriocins.
In biofilm, certain bacteria express specific or non-specific proteins (bacteriocins) which can affect other bacteria. Bacteriocins are different from traditional antibiotics in that they usually only act on its own strain or strains similar to the producing species.Citation36–Citation39
Studies have shown that Streptococci possess the strongest ability of producing bacteriocins among all oral bacteria.Citation39 Bacteriocins produced by S. mutans can be differentiated into two types termed mutacins. The first type are lantibiotics. Lantibiotics are a class of antibiotic peptides containing a characteristic polycyclic thioether, the amino acids lanthionine or methyllanthionine, as well as the unsaturated amino acids dehydroalanine and 2-aminoisobutyric acid. The lantibiotics are synthesized by many Gram positive bacterial species. Lantibiotics contain mutacins I, II and III. The second type are non-lantibiotics containing mutacins IV and V. The inhibition spectrum of lantibiotics on Grampositive bacteria is relatively wide, while the non-lantibiotics have a narrow antimicrobial spectrum, only acting on S. sanguinis.Citation37 Mutacins can enhance the competitiveness of S. mutans successful colonization and its ability to flourish in the biofilm.Citation39,Citation40 In addition, Streptococcus salivarius is an important oral probiotic, and its production of bacteriocins, such as salivaricin A (SalA), strongly inhibits Streptococcus pyogenes.Citation38 Therefore, the bacteriocins expressed by some strains greatly affect other strains living in biofilm.
Recently, Perry et al. found that when the concentration of competence-stimulating peptide (CSP; see below) is high in the micro-environment, mutacin V can be used as a precursor of intracellular auto-active bacteriocin or death effector, mediating S. mutans autolysis via a novel mechanism, which is vividly called intracellular action against self. Perry speculated that there may be some relationship between the autolysis of S. mutans and genetic transformation in the bacterium.
Bacteriocins have a broad prospect for application in the field of medicine. A new class of drugs can be developed which is targeted and contains only minor side effects, using its unique narrow- spectrum antibacterial properties, to avoid microbial flora and bacterial resistance and achieve the desired clinical effect under the condition of ensuring the balance of normal flora in the human body.
Quorum sensing (QS).
Quorum sensing is the self-induced secretion of one or more agents (autoinducer, AI) in response to changes in bacterial density and the surrounding environment, i.e., the bacteria reach a certain threshold (or quorum of cell types, the number of colonies or bacteria), which initiates gene expression to regulate cell or group behavior. Different groups of bacteria have different effects of conditioning; many bacteria secrete the same inducer, thereby regulating the interaction between different types of bacterial behavior.
In biofilm, the process of bacteria producing signal molecules, transporting, sensing and controlling a series of acts is called a quorum sensing system. The majority of biofilm bacteria can sense the bacterial biofilm microenvironment through these systems to control their own physiological characteristics, however, planktonic bacteria do not exhibit these physiological functions.Citation43,Citation44 QS systems control a wide range of responses, including bacterial surface adhesion, extracellular matrix production, synthesis of biosurfactants, spore formation, competency, bioluminescence and virulence factor expression, etc.Citation45,Citation46 QS systems are highly specific and accurate, which are the basis of precise regulations of the different bacterial phenotypes. In complex microbial communities, this careful regulation is truly fascinating because each bacterium can also produce a variety of QS signaling molecules with a variety of signal systems (such as the Vibrio harveyi signaling system with three kinds of AI) but those signaling molecules mainly regulate this particular species instead of affecting other species.Citation47,Citation48 Different signaling molecules in various signaling pathways can also influence each other, constituting a sophisticated three-dimensional network of multi-level and large-scale signaling systems.Citation49
There are three types of recognized QS systems in bacteria: the first type is in Gram-positive bacteria and uses oligopeptides as signal molecules; the second type in Gram-negative bacteria uses an acyl-homoserine lactone as a signal molecule; and the third type is conserved in many Gram-negative and Gram-positive bacteria using AI-2 as a signal molecule.Citation50
Oligo-peptides do not enter the cell. They usually bind to membrane sensor proteins of the two-component signal transduction system, but some of them are usually transported to the cytoplasm before binding to their respective receptors.Citation51,Citation52 Grampositive bacteria such as Staphylococcus aureus often use this complex QS system in which modied oligo-peptides serve as AIs.Citation53
The acyl homoserine lactones (AHL) are usually detected through binding or activating cytoplasmic receptor proteins, which dimerize when the AHL binds. The receptor proteins can also bind to promoter regions of target genes, and then activate or repress the transcription of genes.Citation54 AHL are produced and recognized uniquely. If one species produces an AHL, this special AHL usually can only be recognized by the same species.Citation53 AHL-type AIs are detected by a sensor kinase, which is a membrane-bound two-component hybrid protein. The phosphoryl group is transferred from a sensor kinase to a histidine phosphotransfer protein and subsequently to a response regulator molecule. In addition, to the AHL-type AI, which is species-specic, an interspecies AI called AI-2 (a furonosyl borate ester) has been reported as a signal ligand for certain QS systems.Citation53
AI-2 is an important signal molecule in multi-species biofilms. AI-2 plays a decisive promoting role in biofilm formation and maturation. The luxS gene expressing AI-2 is conserved among many species of bacteria,Citation55 including S. mutans, Streptococcus gordonii, Streptococcus oralis, P. gingivalis, A. actinomycetemcomitans and other oral microorganisms.Citation56,Citation57 AI-2 can also mediate oral Streptococci and Actinomycetes to form mixed biofilm.Citation58 The formation of biofilm depends on AI-2 produced by oral Streptococci. For example, bacteria expressing a mutation in the luxS gene in oral Streptococci cannot express AI-2, resulting in a failure of communication between oral Streptococci and Actinomycetes. This failure leads to loose biofilm formation and significantly decreased bacterial density.Citation51 In recent years, reports of interference of these bacterial QS systems and new ways of inhibiting biofilm formation, has opened new avenues for intractable infectious disease research.
Competence-stimulating peptide (CSP).
QS, which is peptide based, has been shown to regulate genetic competence,Citation59 biofilm formation,Citation60 and acid toleranceCitation61 of bacteria. Recent evidence has shown that competence-stimulating peptide (CSP) can induce alarmones which are intracellular signal molecules and are produced due to harsh environmental factors, and can convey sophisticated messages in a population including the induction of altruistic cellular suicide under stressful conditions. CSP is a small molecule which induces bacteria into competence and then the bacteria can obtain exogenous DNA and change genetic information.Citation62
S. mutans usually grows on enamel surfaces in biofilms and it is often considered as the major etiological agent of dental caries in humans.Citation47Candida albicans is also commonly found in human dental caries, where S. mutans interacts with C. albicans. S. mutans inhibits C. albicans ability to produce germ tubes in cocultures even when S. mutans is physically separated from C. albicans. A recent study shows that the QS molecule CSP, secreted during S. mutans early growth stages, inhibits formation of C. albicans germ tubes.Citation63
In S. mutans, the QS systems consist of approximately five proteins, including a 21-amino-acid CSP. It has been reported that the comC gene encodes CSP, CSP is an inactive precursor at that point, but it is then cleaved and exported by the ABC transporter, ComAB.Citation64 In S. mutans, genetic competence and bacteriocin production are regulated by the ComED system which specifically senses and responds to CSP.Citation65 CSP is synthesized in the cell and released into the extracellular medium, mediating the QS system. When the bacterial density of biofilm increases, CSP molecules in the external environment reach a certain numerical threshold concentration and begin to regulate bacterial density. CSP can regulate physiological activities of S. mutans, including biofilm formation, generation of bacteriocins, stress response, acid tolerance and genetic conversion, etc.Citation37,Citation44 Previous studies in our lab demonstrated that spaP is regulated by CSP because the spaP gene was abolished in com gene-deficient strains. However, direct addition of synthetic CSP to a low-density culture failed to repress spaP gene expression (Chen and Gregory, personal observation).
CSP signaling molecules are highly species-specific. A CSP produced by one bacterium rarely interferes with the activity of CSP produced by a different bacterium. Eckert et al. has developed a specific highly novel antibacterial agent using the species specific nature of CSP, called specifically targeted antimicrobial peptides (STAMPs). STAMPs contain two functional domains, the first domain is a targeting peptide domain and the second domain is an antimicrobial peptide domain. The antibacterial domain recognizes S. mutans and kills it, but there is no effect on other types of non-cariogenic streptococci. This confirms the specificity of the STAMP.
Bacterial Species Interactions in Biofilms
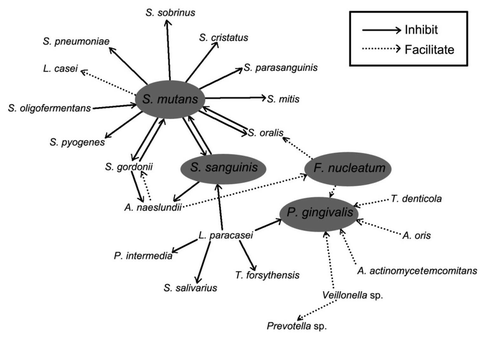
Due to limited growing space and nutrients, bacteria from different species compete with each other in biofilms (). Every species has it own stratagem to compete against others.
Streptococcus mutans.
S. mutans is considered as one of the most important cariogenic bacteria.Citation67 It is a common bacterial species in human beings because it is present in almost everyone's oral cavity. However, in caries-free oral cavities the percentage of S. mutans is very low; while in caries-affected oral cavities the percentage of S. mutans is high. The shift of S. mutans proportion from low percentage to high percentage is associated with caries.Citation68
Kreth et al.Citation19 reported dual species inhibitory activities of S. mutans on S. gordonii, S. pyogenes, Streptococcus mitis ATCC 903, Streptococcus pneumoniae, Streptococcus cristatus and S. sanguinis were significant; the inhibitions on S. oralis, S. mitis ATCC 33399 and S. parasanguinis were medium; and the inhibition on S. sobrinus was mild.
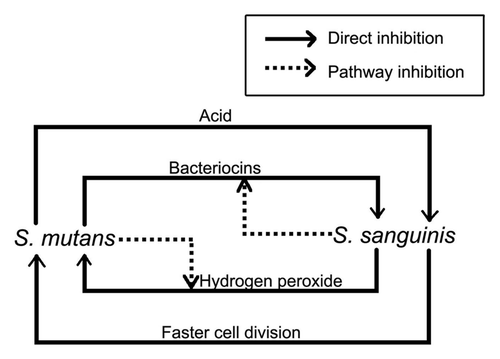
The numbers of S. mutans and S. sanguinis are negatively associated. If the level of S. mutans is high, the level of S. sanguinis is low; while if the level of S. sanguinis is high, the level of S. mutans is low. This possibly explains the reason why cariesactive individuals have low numbers of S. sanguinis and caries-free subjects have higher numbers of S. sanguinis. In a dual-species S. mutans and S. sanguinis biofilm, if S. mutans colonized first, the growth of the later colonized S. sanguinis would be inhibited by S. mutans. However, if S. sanguinis colonized first, the growth of the later colonized S. mutans would be inhibited by S. sanguinis. Furthermore, if S. mutans and S. sanguinis colonized together at the same time, either species can inhibit the other. Therefore, the order of colonization determines the result of the competition.Citation19 This result confirmed earlier research done by Kemp et al. in 1983.Citation69 Clinical studies also confirmed the competition between S. gordonii and S. mutans or between S. sanguinis and S. mutans.Citation70 Further researchCitation19 suggested the inhibition of S. mutans on S. sanguinis was caused by an excreted diffusible substance instead of nutrient deprivation. However, the amount of nutrients present plays a significant role in the competition. When nutrients were rich, both S. mutans and S. sanguinis grew well, no matter which species colonized first.
One mechanism of S. mutans inhibition on S. sanguinis is the large amount of organic acid S. mutans produces. Although both S. mutans and S. sanguinis can metabolize glucose and produce lactate when incubated with excess glucose, S. mutans produces more acid than S. sanguinis due to the higher ATP-glucose phosphotransferase activity of S. mutans than S. sanguinis.Citation71 Therefore, the lactic acid produced by S. mutans glucose metabolism is excreted into the environment and inhibits the growth of S. sanguinis, resulting in an increased ratio of S. mutans over S. sanguinis.Citation72 Kreth et al.Citation19 confirmed these results. When S. mutans and S. sanguinis were cultivated under acidic conditions (pH 5.5) at the same time, S. mutans grew better than S. sanguinis. Acidic conditions can repress or damage the ATP-glucose phosphotransferase activity of both strains, but the repression on S. mutans was less than on S. sanguinis ().Citation72
A second mechanism of S. mutans inhibition on S. sanguinis is the mutacin S. mutans excretes. Both S. mutans mutacins I and IV can inhibit the growth of S. sanguinis.Citation65 The mutacin inhibition order of S. mutans on S. sanguinis from the smallest to the largest was double-mutant strain (I− IV−), mutacin I-defective strain (I− IV+), mutacin IV-defective strain (I+ IV−) and wild-type strain (I+ IV+). The presence or absence of oxygen affects mutacin expression. Under aerobic conditions the mutacin IV structural gene was upregulated 5-fold compared with a non-anaerobic condition. But no significant difference was observed in mutacin I gene expression between the two oxygen conditions ().Citation40
A third mechanism is that S. mutans inhibits the ability of S. sanguinis to produce hydrogen peroxide (H2O2). The H2O2 production of S. sanguinis was 66% reduced when S. sanguinis was cultivated with S. mutans compared with cultivated alone ().Citation19
Streptococcus sanguinis.
S. sanguinis is one of the early colonizers in biofilm and is considered a beneficial species in the oral biofilm ecosystem. It can produce H2O2 as a means of excreting excessive oxygen and serves as a non-specific antimicrobial agent, which can inhibit S. mutans and other anaerobic periodontal pathogens growth.Citation73 However, it is obtrusive to say S. sanguinis is 100% benign due to its association with bacterial endocarditis.Citation74
S. sanguinis competes with other bacteria by producing a bacteriocin of S. sanguinis (sanguicin) or excreting H2O2.Citation75 H2O2 can inhibit the glycolysis and protein synthesis of target bacteria. Baldeck and MarquisCitation76 reported both biofilm and planktonic S. mutans cells were sensitive to H2O2. Low concentrations (16.3 mmol/L) of H2O2 could strongly inhibit glycolysis and are primarily bacteriostatic. The most sensitive target was glyceraldehyde-3-phosphate dehydrogenase and a less sensitive target was the phosphoenolpyruvate:glucose phosphotransferase pathway. High concentrations (30 mmol/L) of H2O2 could inhibit protein synthesis, resulting in cell damage due to methionine oxidation or lack of cell repair mechanisms.Citation76,Citation77 A eukaryotic-type serine/threonine protein kinase (STPK) in the S. mutans cell-envelope is related to H2O2 antimicrobial resistance and a STPK-defective strain of S. mutans was observed to be more sensitive to H2O2.Citation78 Kreth et al.Citation40 observed the inhibition of S. mutans by S. sanguinis was reduced when grown with glucose. The reason of this reduction in inhibition is that glucose may repress the expression of pyruvate oxidase. Pyruvate oxidase is related to H2O2 formation, as H2O2, acetyl phosphate and carbon dioxide are synthesized from pyruvate by pyruvate oxidase. Therefore, glucose indirectly represses the excretion of H2O2 by S. sanguinis. The amount of oxygen in the environment also affects the competition between S. sanguinis and S. mutans. The inhibition of S. mutans by S. sanguinis is increased under aerobic conditions compared with anaerobic conditions.Citation40S. sanguinis inhibits S. mutans production of mutacin. The mutacin production of S. mutans was 20% of when cultivated with S. sanguinis compared with cultivated alone.Citation65 Furthermore, the doubling time of S. sanguinis is shorter than the generation time of S. mutans, and this more rapid reproduction facilitates S. sanguinis competing against S. mutans.Citation79
Previously, we investigated the secretion of immunoglobulin A1 (IgA1) protease of S. sanguinis in dual-species culture with S. mutans. We found the IgA1 protease of S. sanguinis was enhanced when it was incubated with S. mutans or Actinomyces viscosus. The enhancement factors from S. mutans or A. viscosus are soluble and less than 14 kDa in size. Further research confirmed that S. mutans strain MT8148, TH16, ATCC 10449, KPSK2, V310, V318, Ingbritt and ATCC 25175, and S. sobrinus strain ATCC 27352 also enhanced IgA1 protease expression of S. sanguinis. These results suggested that the enhancing factors may serve an ecological role facilitating S. mutans, A. viscosus and S. sobrinus to evade anti-bacterial IgA antibodies in saliva and gain an opportunity to multiply.Citation80
Streptococcus gordonii.
S. gordonii is one of the pioneer bacteria attached to supragingival pellicle. Clinical studies confirmed competition between S. gordonii and S. mutans.Citation70 Kreth et al.Citation40 observed the inhibition of S. mutans by S. gordonii, but this inhibition was less than the inhibition of S. mutans by S. sanguinis. Like S. sanguinis, the inhibition of S. mutans by S. gordonii is stronger under aerobic conditions than under anaerobic conditions.Citation40 Interestingly, the inhibition was reduced with glucose. The mechanism of this reduction is similar to that of S. sanguinis, in that pyruvate oxidase is affected and the amount of H2O2 is reduced. This decrease in inhibition was more significant in S. gordonii than in S. sanguinis.Citation40
Furthermore, S. gordonii inhibits A. naeslundii growth by excreting H2O2. S. gordonii is usually detected together in dental biofilm with Veillonella species. Additional research reported S. gordonii can coaggregate with Veillonella species.Citation28
Streptococcus oralis.
The doubling time of S. oralis is shorter that the generation time of S. mutans, and this more rapid reproduction facilitates S. oralis competing against S. mutans.Citation79 S. oralis is typically detected together with Veillonella species, while additional research reported S. oralis can coaggregate with Veillonella species.Citation28
Streptococcus oligofermentans.
S. oligofermentans can produce H2O2 mainly by two methods. The first method is to metabolize the lactic acid produced by S. mutans into H2O2 by lactate oxidase, while the second method is to metabolize the peroxide to H2O2 by L-amino acid oxidase. The H2O2 produced by S. oligofermentans can cooperate with the H2O2 produced by S. sanguinis and increase inhibition of S. mutans growth.Citation73
Porphyromonas gingivalis.
In a series of dual-species experiments, P. gingivalis was demonstrated to grow with S. gordonii, Actinomyces oris, Veillonella sp, F. nucleatum and A. actinomycetemcomitans, but not with S. oralis. In a three-species model,Citation81 P. gingivalis was demonstrated to grow with dual-species of Veillonella sp and A. actinomycetemcomitans, and dual-species of Veillonella sp and F. nucleatum, but P. gingivalis does not grow with either dual-species of S. oralis and A. actinomycetemcomitans or dual-species of S. oralis and F. nucleatum. Therefore, P. gingivalis apparently cannot grow in the presence of S. oralis. However, in a three-species biofilm with S. gordonii and S. oralis, P. gingivalis grew well,Citation81 this result indicated that S. gordonii facilitated P. gingivalis growth.
P. gingivalis is usually detected together with Treponema denticola in subgingival plaque. T. denticola can produce succinic acid which was identified in the phospholipids and lipids on the P. gingivalis cell envelope.Citation82 However, P. gingivalis produces isobutyric acid which increases T. denticola growth.Citation83 Therefore, a cooperative synergistic growth relationship may exist between P. gingivalis and T. denticola.
Fusobacterium nucleatum.
Fusobacteria are the predominate Gram-negative bacterial species in mature dental plaque. However, in the initial colonization, streptococci and actinomyces are the predominate species. The coaggregation of Fusobacteria with other bacterial species is a pivotal link between primary and secondary dental plaque colonization.Citation20,Citation84 F. nucleatum is a crucial microorganism in dental biofilms because of its coordinator role, which coaggregates with both late and early dental biofilm colonizers.Citation20
The coaggregation between F. nucleatum and other Gram-negative bacterial species are governed by lectin-carbohydrate receptor interactions. For example, the coaggregation between P. gingivalis and F. nucleatum is regulated by a galactoside P. gingivalis moiety and a F. nucleatum lectin, which can be inhibited by lactose. P. gingivalis capsular polysaccharides and lipopolysaccharides are involved through receptors which mediate coaggregation between oral bacterial species.Citation85 In contrast, although coaggregation between F. nucleatum and many Gram-positive bacterial species has been observed, those coaggregations are rarely inhibited by sugars.Citation26,Citation86,Citation87 Thus, intergeneric coaggregation between F. nucleatum and Gram-negative organisms is significantly distinct from F. nucleatum binding to Gram-positive bacteria.
Recent research demonstrates that F. nucleatum is difficult to grow in saliva without the presence of A. naeslundii. Interestingly S. oralis cells increased in the presence of F. nucleatum. Therefore, Fusobacteria require A. naeslundii for its own biofilm formation and improve the initial colonization of streptococci in certain dual-species biofilms.Citation88
Oral bacteria will confront oxygen stress in dental biofilm. The survival of oral anaerobic bacterial species depends on the specific tolerance of each species to oxygen and the bacterial interactions within the biofilm community. F. nucleatum supports P. gingivalis growth by providing a capnophilic environment when growing in an oxygenated and CO2 depleted environment.Citation89 In a two stage chemostat system, the survival of obligate anaerobic microorganisms in an aerated environment was improved by the coaggregation with F. nucleatum.Citation28
Lactobacillus casei.
L. casei is commonly detected in cariogenic biofilm. This strain can produce abundant lactic acid and has a wide pH range. However, the tooth surface adherence of this acid tolerant strain is weak. Wen et al. observed that the biofilm formation of L. casei alone was less than the biofilm formation of dual-species culture with S. mutans. The reason for this improvement was that S. mutans provided L. casei glucosyltransferase (GTF) enzymes, with which glucans were synthesized, and therefore bacterial adhesion was improved.
Lactobacillus paracasei.
L. paracasei produces bacteriocins, which make pores in the cytoplasmic membranes of P. gingivalis, P. intermedia, T. forsythensis, S. salivarius and S. sanguinis, inhibiting the growth of those strains. This bacteriocin has a molecular weight of approximately 56 kDa with a composition of 68% carbohydrate and 32% protein. This protein has cationic amino acids present near the N-terminus, hydrophobic amino acids present in the central region and hydrophilic residues present at the C-terminus.Citation90,Citation91
Actinomyces naeslundii.
A. naeslundii is one of the pioneer bacteria in dental biofilm formation, and it is related to periodontal disease and root caries. A. naeslundii can protect many bacterial species from H2O2, because A. naeslundii consumes H2O2 by protein oxidation.Citation92A. naeslundii also upregulates the spxB gene (which encodes for pyruvate oxidase) of S. gordonii. Furthermore, A. naeslundii can provide S. gordonii arginine, which is related to the protein oxidation effect exhibited in A. naeslundii. The residual amount of S. gordonii intracellular arginine is limited due to the excretion of H2O2. Therefore, growth of S. gordonii was increased by aggregation with A. naeslundii. In the presence of A. naeslundii, S. gordonii can grow under arginine depletion conditions in which S. gordonii usually does not survive.Citation93,Citation94
Veillonella.
Veillonella species can produce vitamin K, which can be utilized by Prevotella spp and Porphyromonas spp for their growth.Citation95Veillonella species utilize the organic acids produced by S. mutans in a symbiotic manner that benefits the growth of both species.
Conclusions
For hundreds of years, scientists and dentists have been looking for efficient methods to control oral diseases. Since the oral ecosystem view has become more widely accepted and oral bacterial interactions have been more elucidated, in the future there may be medications to take advantage of inter-bacterial antagonism to control and prevent those diseases. However, future research must address the interactions within biofilm which are not fully understood today. Because of the complex and variable nature of biofilm, this goal can only be fulfilled with future investigations focused on the model of oral ecosystems and relative factors, with advanced understanding of the mechanisms of bacterial biofilm interaction mechanisms.
Figures and Tables
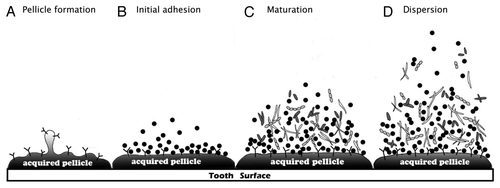
Figure 1 Oral biofilm formation. This diagram represents each step of oral biofilm formation. (A) Pellicle formation. The pellicle is a thin film derived from salivary glycoproteins attached to a clean tooth surface. (B) Initial adhesion. Pioneer bacteria in saliva recognize the binding proteins in acquired pellicle and attach to them. This adhesion is reversible. (C) Maturation. Different bacterial species coaggregate and mature biofilm forms. (D) Dispersion. Bacteria disperse from the biofilm surface and spread to colonize a new site.
Acknowledgments
This work was partially funded by NIH grants HL098960 and DE020614, the Indiana University Purdue University Indianapolis Tobacco Cessation and Biobehavioral Group, and the Indiana University School of Dentistry Ph.D. Student Research Fund.
References
- Dobell C. The rst observations on entozoic protozoa and bacteria. Antony Van Leeuwenhoek and His ‘Little Animals’ 1958; New York Russell and Russell, Inc 236 - 256
- Marsh PD. Dental plaque: biological signicance of a biolm and community life-style. Journal of Clinical Periodontology 2005; 32:7 - 15
- Madigan MT, Martinko JM. Brock Biology of microorganisms 2006; 617:Upper Saddle River Prentice Hall, Inc 618
- Kolenbrander PE, Palmer RJ Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000; 42:47 - 79
- Hojo K, Nagaoka S, Ohshima T, Maeda N. Bacterial interactions in dental biofilm development. J Dent Res 2009; 88:982 - 990
- Geesey GG, Lewandowski Z, Flemming HC. Biofouling and Biocorrosion in Industrial Water Systems 1994; Ann Arbor Lewis Publishers
- Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 2003; 50:61 - 68
- Kenneth J, Ryan C, Ray G. Sherris' medical microbiology [M] 2004; 4:New York McGraw Hill Medical Pub Division 141 - 149
- Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA 2005; 102:7952 - 7957
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000 2006; 42:80 - 87
- Roberts FA, Darveau RP. Beneficial bacteria of the periodontium. Periodontol 2002; 30:40 - 50
- Hannig C, Hannig M. The oral cavity—a key system to understand substratum-dependent bioadhesion on solid surfaces in man. Clinical Oral Investigations 2009; 13:123 - 139
- Gray JJ. The interaction of proteins with solid surfaces. Current Opinion in Structural Biololy 2004; 14:110 - 115
- Characklis WG, Marshall KC. Biofilms 1990; New York John Wiley & Sons, Inc
- Handley PS. Structure, composition and functions of surface structures on oral bacteria. Biofouling 1990; 2:239 - 264
- Ritz HL. Microbial population shifts in developing human dental plaque. Archives of Oral Biologh 1967; 12:1561 - 1568
- Foster JS, Kolenbrander PE. Development of a multi-species oral bacterial community in a saliva-conditioned flow cell. Appl Environ Microbiol 2004; 70:4340 - 4348
- Dige I, Nyengaard JR, Kilian M, Nyvad B. Application of stereological principles for quantification of bacteria in intact dental biofilms. Oral Microbiol Immunol 2009; 24:69 - 75
- Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 2005; 187:7193 - 7203
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ. Communication among oral bacteria. Microbiology and Molecular Biology Reviews 2002; 66:486 - 505
- Kolenbrander PE, London J. Ecological signicance of coaggregation among oral bacteria. Advances in Microbial Ecology 1992; 12:183 - 217
- McIntire FC, Vatter AE, Baros J, Arnold J. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infection and Immunity 1978; 21:978 - 988
- Kaplan JB. Biofilm dispersal: mechanisms, clinical implications and potential therapeutic uses. J Dent Res 2010; 89:205 - 218
- Sanui T, Gregory RL. Analysis of Streptococcus mutans biofilm proteins recognized by salivary immunoglobulin A. Oral Microbiol Immunol 2009; 24:361 - 368
- Roberts SK, Bass C, Brading M, Lappin-Scott H, Stoodley P. New-man HN, Wilson M. Biolm information and structure: what's new?. Dental plaque revisited 1999; Cardiff Bioline 15 - 35
- Kolenbrander PE, Andersen RN, Moore LV. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun 1989; 57:3194 - 3203
- Metzger Z, Blasbalg J, Dotan M, Weiss EI. Enhanced attachment of Porphyromonas gingivalis to human fibroblasts mediated by Fusobacterium nucleatum. J Endod 2009; 35:82 - 85
- Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun 1998; 66:4729 - 4732
- Takahashi N. Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol 2003; 18:109 - 113
- Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 2002; 292:107 - 113
- McCann KS. The diversity-stability debate. Nature 2000; 405:228 - 233
- Costerton JW. Introduction to biolm. International Journal of Antimicrobial Agents 1999; 11:217 - 221
- Levy SB. The challenge of antibiotic resistance. Scientific American 1998; 278:46 - 53
- Erickson PR, Herzberg MC. Emergence of antibiotic resistant Streptococcus sanguis in dental plaque of children after frequent antibiotic therapy. Pediatric Dentistry 1999; 21:181 - 185
- Sedlacek MJ, Walker C. Antibiotic resistance in an in vitro subgingival biofilm model. Oral Microbiol Immunol 2007; 22:333 - 339
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev 2007; 71:653 - 670
- Senadheera D, Cvitkovitch DG. Quorum sensing and biofilm formation by Streptococcus mutans. Adv Exp Med Biol 2008; 631:178 - 188
- Chatterjee C, Paul M, Xie L, Wilfred A, Donk WA. Biosynthesis and mode of action of lantibiotics. Chem Rev 2005; 105:633 - 684
- Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol 2007; 189:1189 - 1198
- Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 2008; 190:4632 - 4640
- Wescombe PA, Upton M, Dierksen KP, Ragland NL, Sivabalan S, Wirawan RE, et al. Production of the lantibiotic salivaricin A and its variants by oral streptococci and use of a specific induction assay to detect their presence in human saliva. Appl Environ Microbiol 2006; 72:1459 - 1466
- Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 2009; 72:905 - 917
- Reading NC, Sperandio V. Quorum sensing: the many languages of bacteria. FEMS Microbiol Lett 2006; 254:1 - 11
- Zhang K, Ou M, Wang W, Ling J. Effects of quorum sensing on cell viability in Streptococcus mutans biofilm formation. Biochem Biophys Res Commun 2009; 379:933 - 938
- Nadell CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biol 2008; 6:171 - 179
- Sifri CD. Healthcare epidemiology: quorum sensing: bacteria talk sense. Clin Infect Dis 2008; 47:1070 - 1076
- Kolenbrander PE. Oral microbial communities: biofilms, interactions and genetic systems. Annu Rev Microbiol 2000; 54:413 - 437
- Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol 2004; 186:6902 - 6914
- Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits and implications for biofilms and disease. Annu Rev Biomed Eng 2008; 10:145 - 167
- Karatan E, Watnick P. Signals, regulatory networks and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 2009; 73:310 - 347
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet 2008; 42:541 - 564
- Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci 2003; 8:32 - 45
- Henke JM, Bassler BL. Bacterial social engagements. Trends Cell Biol 2004; 14:648 - 656
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 1994; 176:269 - 275
- Bassler BL. Small talk: Cell-to-cell communication in bacteria. Cell 2002; 109:421 - 424
- Xie H, Lin X, Wang BYWJ, Lamont RJ. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 2007; 153:3228 - 3234
- James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun 2006; 74:3834 - 3844
- Rickard AH Jr, Palmer RJ, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol 2006; 60:1446 - 1456
- Li Y, Hanna MN, Svensater G, Ellen RP, Cvitkovitch DG. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J Bacteriol 2001; 183:6875 - 6884
- Li Y, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 2001; 183:897 - 908
- Li Y, Tang N, Aspiras MB, Lau PCY, Lee JH, Ellen RP, Cvitkovitch DG. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 2002; 184:2699 - 2708
- Petersen FC, Pecharki D, Scheie AA. Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J Bacteriol 2004; 186:6327 - 6331
- Jarosz LM, Deng D, van der Mei Henny C, Crielarrd W, Krom BP. Streptococcus mutans Competence-Stimulating Peptide Inhibits Candida albicans Hypha Formation. Eukaryotic Cell 2009; 8:1658 - 1664
- Petersen FC, Scheie AA. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol Immunol 2000; 15:329 - 334
- Kreth J, Merritt J, Shi W, Qi F. Coordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighboring species. Mol Microbiol 2005; 57:392 - 404
- Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob Agents Chemother 2006; 50:3651 - 3657
- Balakrishnan M, Simmonds RS, Tagg JR. Dental caries is a preventable infectious disease. Australian Dental Journal 2000; 45:235 - 245
- Marsh PD. Are dental diseases examples of ecological catastrophes?. Microbiology 2003; 149:279 - 294
- Kemp CW, Robrish SA, Curtis MA, Sharer SA, Bowen WH. Application of a competition model to the growth of Streptococcus mutans and Streptococcus sanguis in binary continuous culture. Appl Environ Microbiol 1983; 45:1277 - 1282
- Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 2002; 40:1001 - 1009
- Komiyama K, Kleinberg I. Comparison of glucose utilization and acid formation by Streptococcus mutans and Streptococcus sanguis at different pH. J dent Res 1974; 53:241
- Iwami Y, Yamada T. Rate-limiting steps of the glycolytic pathway in the oral bacteria Streptococcus mutans and Streptococcus sanguis and the influence of acidic pH on the glucose metabolism. Arch Oral Biol 1980; 25:163 - 169
- Tong H, Chen W, Shi W, Qi F, Dong X. SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J Bacteriol 2008; 190:4716 - 4721
- Stinson MW, Alder S, Kumar S. Invasion and killing of human endothelial cells by viridans group streptococci. Infect Immun 2003; 71:2365 - 2372
- Merritt J, Kreth J, Shi W, Qi F. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol Microbiol 2005; 57:960 - 969
- Baldeck JD, Marquis RE. Targets for hydrogen-peroxide-induced damage to suspension and biolm cells of Streptococcus mutans. Can J Microbiol 2008; 54:868 - 875
- Skaar EP, Tobiason DM, Quick J, Judd RC, Weissbach H, Etienne F, et al. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc Natl Acad Sci USA 2002; 99:10108 - 10113
- Zhu L, Kreth J. Role of Streptococcus mutans eukaryotic-type serine/threonine protein kinase in interspecies interactions with Streptococcus sanguinis. Arch Oral Biol 2010; 55:385 - 390
- Wen ZT, Yates D, Ahn SJ, Burne RA. Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol 2010; 10:111
- Gregory RL, Lloyd DR. Enhancement of Streptococcus sanguis immunoglobulin A1 protease activity by Streptococcus mutans. Immunol Infect Dis 1993; 3:309 - 315
- Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early and late colonizers of enamel. J Bacteriol 2009; 191:6804 - 6811
- Lev M. Sphingolipid biosynthesis and vitamin K metabolism in Bacteroides melaninogenicus. Am J Clin Nutr 1979; 32:179 - 186
- Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun 1992; 60:5298 - 5301
- Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol 1993; 175:3247 - 3252
- Rosen G, Sela MN. Coaggregation of Porphyromonas gingivalis and Fusobacterium nucleatum PK 1594 is mediated by capsular polysaccharide and lipopolysaccharide. FEMS Microbiol Lett 2006; 256:304 - 310
- Kang MS, Na HS, Oh JS. Coaggregation ability of Weissella cibaria isolates with Fusobacterium nucleatum and their adhesiveness to epithelial cells. FEMS Microbiol Lett 2005; 253:323 - 329
- Nagaoka S, Hojo K, Murata S, Mori T, Ohshima T, Maeda N. Interactions between salivary Bifidobacterium adolescentis and other oral bacteria: in vitro coaggregation and coadhesion assays. FEMS Microbiol Lett 2008; 281:183 - 189
- Periasamy S, Chalmers NI, Du-Thumm L, Kolenbrander PE. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl Environ Microbiol 2009; 75:3250 - 3257
- Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbondioxide-depleted environments. Microbiology 2002; 148:467 - 472
- Pangsomboon K, Kaewnopparat S, Pitakpornpreecha T, Srichana T. Antibacterial activity of a bacteriocin from Lactobacillus paracasei HL32 against Porphyromonas gingivalis. Arch Oral Biol 2006; 51:784 - 793
- Pangsomboon K, Bansal S, Martin GP, Suntinanalert P, Kaewnopparat S, Srichana T. Further characterization of a bacteriocin produced by Lactobacillus paracasei HL32. J Appl Microbiol 2009; 106:1928 - 1940
- Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 2003; 25:207 - 218
- Jakubovics NS, Gill SR, Iobst SE, Vickerman MM, Kolenbrander PE. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol 2008; 190:3646 - 3657
- Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol 2008; 66:637 - 644
- Marcotte H, Lavoie MC. Oral microbial ecology and the role of salivary immunoglobulin A. Microbiol Mol Biol Rev 1998; 62:71 - 109