Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is abundant in hospitals and in the United States is a leading cause of mortality due to infectious agents. Community-associated MRSA (CA-MRSA) strains such as USA300, which typically cause disease outside of healthcare settings, are also prevalent in the United States. Although most CA-MRSA infections affect skin and soft tissue, the pathogen can enter the bloodstream and ultimately cause severe disease. In a recent paper, we used USA300-specific microarrays to generate a comprehensive view of the molecules that facilitate S. aureus immune evasion and survival in human blood. Notably, genes encoding proteins involved in iron-uptake and utilization and gamma-hemolysin (hlgABC) are highly up-regulated by USA300 during culture in human blood. Here we discuss the potential implication of these findings and the possible role of gamma-hemolysin in the success of S. aureus as a human pathogen.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading cause of nosocomial infections worldwide and endemic in hospitals of most industrialized countries. In addition, MRSA is now a prominent cause of infection outside of hospital settings. These so-called community-associated MRSA (CA-MRSA) infections are widespread in many regions of the world, including the United States. A strain known as USA300 is the most prevalent cause of CA-MRSA infections in the United States and in 2004 was reported as the most abundant cause of all bacterial skin infections in the community.
The vast majority of CA-MRSA infections are those that affect skin and soft tissue. Nevertheless, bacteria can gain entry into the bloodstream directly through a break in the skin or from some other anatomical site of infection. A nationwide surveillance study performed in the United States (1995–2002) showed that S. aureus was responsible for approximately 20% of all bloodstream infections, and 20–30% of those were fatal. Although significant progress has been made, the mechanisms that allow S. aureus—especially CA-MRSA strains such as USA300—to survive in human blood and ultimately cause serious disease remain incompletely characterized.
In a recent study, we used custom USA300-specific Affymetrix microarrays to analyze changes in the USA300 transcriptome that occur during short-term (up to 2 h) culture in human blood. The in vitro model system was designed to mimic conditions imparted on S. aureus immediately after the pathogen enters the bloodstream. Culture of USA300 in blood triggered changes in the expression of genes encoding molecules involved in amino acid transport and metabolism, coenzyme transport and metabolism, inorganic ion transport and metabolism, nucleotide transport and metabolism, and secondary metabolite biosynthesis. Notably, genes encoding a vast repertoire of proteins involved in iron uptake and utilization, including the iron-regulated surface determinants (Isd) of S. aureus, proteins involved in siderophore mediated transport (Sir family), and siderophore biosynthesis proteins, were upregulated.
The concentration of free iron in human blood that is easily accessible for bacteria is ∼1011 times lower than that required for bacterial survival. Inasmuch as S. aureus has had a long-term relationship with humans as both a commensal organism and pathogen, it co-evolved to acquire free iron as well as that bound to transferrin or blood hemoproteins. The requirement for iron is reflected by changes in the S. aureus transcriptome following culture in whole blood or human serum.
To survive in human blood, S. aureus must employ multiple mechanisms to avoid killing by the innate immune system. The expression of ∼25% of all S. aureus genes encoding defense mechanisms and known and putative virulence factors is altered in human blood, and 15% of these genes are upregulated. Among the genes upregulated are those encoding intercellular adhesion protein B (icaB), extracellular fibrinogen-binding protein (efb), secretory extracellular matrix and plasma binding protein (empbp), immunoglobulin G binding protein A precursor (spa), murein hydrolase regulator subunits A and B (lrgA and lrgB), and superantigen-like proteins and leukotoxins, such as leukotoxin GH (LukGH), Panton-Valentine leukocidin (PVL) and gamma-hemolysin (Hlg).
Along with LukGH and PVL, Hlg belongs to the family of synergohymenotropic (bi-component) pore-forming toxins, although each has somewhat unique specificity for host cells. Unlike other bi-component toxins that have one S and one F subunit, Hlg contains two S subunits (HlgA and HlgC) and one F subunit (HlgB), which can be assembled into HlgAB or HlgBC pairs. Only HlgAB displays high hemolytic activity toward both rabbit and human erythrocytes and also functions as a leukotoxin.
Does Hlg Promote S. aureus Survival in Human Blood?
From a mechanism standpoint, HlgA was shown to be the subunit that binds directly to the erythrocyte membrane. The paucity of free iron in human blood coupled with the proven high hemolytic activity of HlgAB likely explains our finding that there was high upregulation of hlgA during culture of S. aureus in human blood. In human blood, hlgA transcript was upregulated 145-fold after 90 min of culture and hlgB and hlgC were upregulated ∼34-fold. Interestingly, when USA300 was cultured in normal human serum—conditions in which leukocytes and erythrocytes are absent—hlgA transcript was minimally upregulated (3-fold) and hlgB and hlgC were downregulated. These findings indicate that interaction with blood cells is important for upregulation of genes encoding Hlg subunits, a notion supported by previous data with purified human neutrophils.
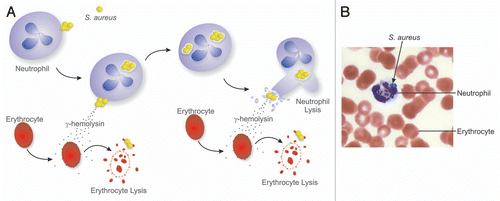
Previous in vivo data suggest Hlg plays a rather modest role in the overall virulence of S. aureus. Consistent with these previous findings, deletion of genes encoding Hlg subunits A, B and C (hlgA, hlgB and hlgC) in USA300 had no effect on the ability of S. aureus to cause skin disease (abscesses and dermonecrosis) in mice. Nonetheless, a USA300 isogenic hlgABC deletion strain (LACΔhlgABC) had reduced survival in human blood compared to the wild-type strain, albeit the difference in survival was transient. These data were perhaps reflected by the slightly decreased ability of LACΔhlgABC to cause death in a mouse bacteremia model compared with the wild-type strain. Based on these findings and the microarray data, one could envision a model whereby Hlg works in concert with iron uptake and metabolism processes to promote S. aureus survival in blood (). After entering the bloodstream, bacteria interact with leukocytes—primarily neutrophils—and erythrocytes. Neutrophils ingest USA300 rapidly, but a significant number of bacteria survive and ultimately cause host cell lysis. The interaction of S. aureus with blood cells triggers upregulation and secretion of Hlg, and the toxin in turn binds to the membranes of erythrocytes and causes lysis of these host cells. Such a process would give S. aureus access to hemoproteins, thereby providing the pathogen with much needed iron ( and B).
Redundancy of Leukotoxins
The virulence phenotype of S. aureus is the result of many molecules working in concert and some of these molecules appear to have redundant function. For example, USA300 has the capacity to produce at least four different two-component leukotoxins, three of which are upregulated in human blood. Inasmuch as the ability of these toxins to target neutrophils is redundant, expression of these molecules is highly dependent on growth conditions and the host environment. PVL and LukGH have specificity for myeloid cells such as neutrophils. By comparison, Hlg targets myeloid cells and erythrocytes. In addition, leukotoxins can assemble as “mix-and-match” subunits (e.g., HlgA + LukF-PV), thereby potentially enhancing or extending target cell specificity. Thus, redundancy of leukotoxins may facilitate increased S. aureus survival in a broad diversity of host environments, such as skin, soft tissue, lungs or blood.
Concluding Comments
Elucidating the relative contribution of specific bacterial molecules to virulence can be a major challenge. The use of system-biology level approaches such as expression microarrays to identify the molecules involved in the process is an important first step toward a comprehensive understanding of bacterial virulence. Our microarray-based analysis highlighted importance of iron-associated proteins for S. aureus survival in blood. In addition, genes encoding Hlg subunits were highly upregulated during culture of S. aureus in human blood. Despite these findings, we found that the toxin has a limited contribution to virulence in mouse infection models. It is possible that the role of Hlg in our model systems was masked by the function of other staphylococcal leukotoxins or cytolytic molecules. Such redundancy among S. aureus virulence molecules is likely important for fitness of the organism. On the other hand, functional redundancy is problematic for studies directed to understand the contribution of individual factors to virulence and/or disease. Perhaps more importantly, the presence of multiple molecules with the same or similar function creates a challenge for the development of new therapeutics or vaccines, which as a consequence must be directed at multiple S. aureus target molecules. Indeed, some of the current S. aureus vaccine strategies employ this very approach. Whether such a strategy will be effective in humans remains an open question.
Figures and Tables
Acknowledgments
The authors are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank Scott D. Kobayashi, Ph.D. (NIAID) for critical review of the manuscript.
Original Article: Malachowa N, Whitney AR, Kobayashi SD, Sturdevant DE, Kennedy AD, Braughton KR, et al. Global changes in Staphylococcus aureus gene expression in human blood. PLoS ONE 2011; 6:18617; PMID: 21525981; http://dx.doi.org/10.1371/journal.pone.0018617