Abstract
The human commensal fungus Candida albicans can cause not only superficial infections, but also life-threatening disease in immunocompromised individuals. C. albicans can grow in several morphological forms. The ability to switch between different phenotypic forms has been thought to contribute to its virulence. The yeast-filamentous growth transition and white-opaque switching represent two typical morphological switching systems, which have been intensively studied in C. albicans. The interplay between environmental factors and genes determines the morphology of C. albicans. This review focuses on the regulation of phenotypic changes in this pathogenic organism by external environmental cues and internal genes.
Introduction
The yeast Candida albicans is a harmless commensal in the oral cavity, digestive tract and genital region of healthy people, while it also causes superficial infections and life-threatening systemic disease.Citation1 With the increase of immunocompromised individuals due to HIV infection, organ transplantation and application of chemotherapy and indwelling devices, invasive candidiasis has become a serious public health problem in the recent several decades.Citation2 The switching from commensal to pathogenic phase has been widely thought to be associated with the phenotypic plasticity of C. albicans. It can grow in several morphological forms including unicellular yeast-form, elongated hyphae and pseudohyphae.Citation3 In this review, hyphae and pseudohyphae are referred to as filamentous forms or cells. A plethora of external and internal factors regulate the switching between different phenotypes ( and and ).Citation3,Citation4 The external factors include environmental cues, that the pathogen often confronts with during its life cycle and host infection, such as serum, high temperature (37°C), low levels of oxygen, high levels of CO2, poor nutrition conditions and so on.Citation4,Citation5 The internal genetic and epigenetic changes play a key role in the regulation of morphogenesis.Citation3-Citation5 In recent years, great strides have been made in uncovering the underlying mechanisms of the morphologic regulation and the coordination and interplay between environmental factors and genes. Several signal transduction pathways and key transcription factors have been intensively investigated.Citation3-Citation5 Another phenotypic switching system, referred to as white-opaque transition, has attracted increasing research interest in the past decade. The underlying molecular mechanisms have begun to be uncovered. This system was first identified in a clinical strain, WO-1, isolated from a transplant patient with a fatal blood stream infection.Citation6 Although the white-opaque transition system gives distinguishable cellular and colony appearance, both the white and opaque forms are budding cells.Citation7 The two types of cells also differ in virulence, sensitivity to immune cells, expression of a wide variety of genes and mating competence. In this review, I will focus on the molecular mechanisms involved in the regulation of yeast-filamentous growth switching as well as white-opaque transition in C. albicans.
Figure 1. Regulation of filamentous growth in C. albicans by multiple environmental cues and signal transduction pathways. The external inducers may function on the cell surface receptors or enter into the cell and directly bind the filamentous growth regulators. The transcription factors Flo8, Efg1 and Cph1 play a central role in the regulation of phenotypic transitions. Multiple signaling pathways converge on the three regulators. The cAMP/PKA pathway and its downstream regulators Flo8 and Efg1 can also play a negative role in filamentous development under embedded growth conditions. The general transcriptional repressor Tup1 is recruited by DNA-binding proteins Nrg1 and Rfg1 and targets on the promoters of hypha-specific genes.
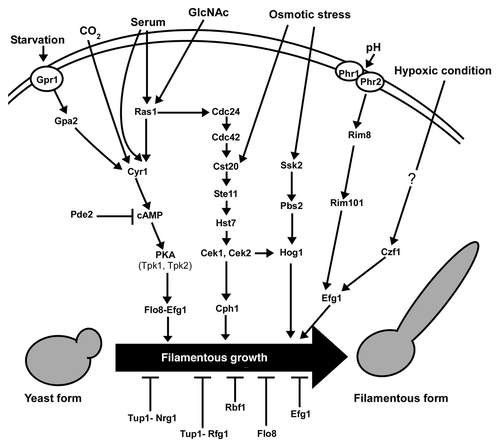
Figure 2. Regulation of white-opaque transition in C. albicans. (A) Environmental factors regulate white-to-opaque and opaque-to-white transitions. (B) The cAMP/PKA pathway and Wor1 involved gene circuitry. The cAMP/PKA pathway regulates both CO2 and GlcNAc induced opaque cell formation. There is also an unidentified pathway mediated CO2 and GlcNAc sensing. The two pathways converge on the master regulator Wor1. The transcription factors Wor1, Wor2, Efg1 and Czf1 form a positive feedback loop controlling white-opaque switching. The inhibition of expression of WOR1 by the MTLa1/α2 heterozygous complex is also shown. The dashed line with an arrowhead represents the unidentified pathway involved in CO2 and GlcNAc sensing.
Table 1. Environmental cues and pathways involved in filamentous growth regulation in C. albicans
Regulation of Yeast-Filamentous Growth Transition
High-frequency phenotypic switching has been observed in C. albicans and other species in the Candida clade.Citation1 In 1985, Soll and colleagues reported that C. albicans can switch between at least seven colony phenotypes.Citation8 The cellular morphologies in this switching system include the unicellular budding yeast and filamentous hyphal and pseudohyphal forms. Yeast-form cells are round and similar to diploid Saccharomyces cerevisiae cells. Hyphae consist of long tubes with no constrictions, while pseudohyphae consist of chains of elongated cells with constrictions between adjacent cells.Citation9 The ability to switch between yeast and filamentous forms is thought to be tightly linked with virulence. Filamentous cells are more invasive and better at tissue penetration, while yeast cells are easy to be delivered and disseminated in the bloodstream.Citation10 In infected tissues, both yeast-form and filamentous cells are found.Citation1 It seems that the ability of switching back and forth between the two forms is important for pathogenesis.
Environmental cues regulate yeast-filamentous transition
It is critical for all organisms to adapt to the changes in their environments. C. albicans can infect almost all human organs.Citation1 It encounters various microenvironmental factors that are unique to the different niches within the host. A range of factors reflecting the host microenvironments have been found to regulate the yeast-filamentous switching. For example, serum is one of the most powerful inducers of filamentous growth in C. albicans. Xu et al. have discovered that bacterial peptidoglycans in the host serum are the major component triggering C. albicans filamentous growth.Citation11 Since the human host cannot synthesize peptidoglycan molecules, it has been indicated that these molecules come from the commensal bacteria in the gastrointestinal tract (GI tract).Citation11 Another two host associated molecules regulating C. albicans yeast-filamentous transition are N-acetylglucosamine (GlcNAc) and CO2.Citation12-Citation16 GlcNAc is a component of the mucus of GI tract and bacterial cell wall, while CO2 is a product of cellular respiration.Citation17,Citation18 The level of CO2 in blood is about 5%, which is much higher than that in ambient atmosphere (0.036%).Citation19 The cAMP/PKA pathway is involved in GlcNAc and CO2 induced filamentous growth.Citation12,Citation13 C. albicans also undergoes morphological changes in response to the host temperature (37°C), neutral pH, nutrient limitation and low O2 levels.Citation3-Citation5,Citation20-Citation22 In vitro experiments demonstrate that the quorum sensing molecule, farnesol and physical interaction (e.g., growth in embedded matrix) regulate morphogenesis in C. albicans.Citation21,Citation23 Interestingly, C. albicans can undergo filamentous growth under embedded conditions at relatively low temperature (25°C).Citation21 During the past two decades, great progress has been made in understanding the underlying mechanisms regulating external factors induced morphogenesis in this fungal pathogen (). The following sections will focus on the major pathways and genes involved in regulation of morphogenesis in response to different environmental factors. Their roles in virulence will also be discussed.
Ras proteins
Ras is a member of a highly conserved family of small GTPases in eukaryotes ranging from yeast to humans.Citation24 Ras is activated when bound with GTP, while it is in an inactivated form when bound with GDP.Citation24 There are two Ras proteins in C. albicans, Ras1 and Ras2.Citation25,Citation26 C. albicans Ras1 is a homolog of S. cerevisiae Ras2, which regulates the downstream MAPK and cAMP/PKA pathways.Citation27 C. albicans Ras2 belongs to a group of atypical Ras proteins and shares poor identity with C. albicans Ras1, S. cerevisiae Ras1 and Ras2.Citation26 Ras1 is required for filamentous growth and virulence in C. albicans. Deletion of RAS1 impairs serum induced hyphal growth, while ectopic expression of a dominant active form RAS1V13 has a promoting effect on hyphal development.Citation25 Supplementing the growth media with cAMP or overexpression of components of the MAPK cascade rescued the filamentous growth defect of the ras1Δ/Δ mutant, suggesting that C. albicans Ras1, like S. cerevisiae Ras2, is upstream of the cAMP and MAPK pathways.Citation27 In a systemic infection model, the ras1Δ/Δ mutant shows notably reduced virulence.Citation25 Ras2 has just been recently characterized in C. albicans. Deletion of RAS2 gene alone in a wild type strain has no notable effect, while disruption of it in a ras1Δ/Δ background mutant results in exacerbated hyphal growth defect. Although recombinant C. albicans Ras2 shows similar GTPase activity as does Ras1, Ras2 has an antagonizing effect on Ras1 at many aspects including regulation of cAMP level, stationary-phase entry and stress response.Citation26 The opposite roles of the two Ras proteins may fine-tune the downstream pathways in respond to different environmental changes.
Cst20-Cst11-Hst7-Cek1/2 mediated MAPK cascade
The roles of the MAPK pathway in morphogenesis and mating have been extensively studied in C. albicans.Citation28-Citation32 The Rho-type GTPase Cdc42 and its exchange factor Cdc24 are required for normal budding, virulence and filamentous growth.Citation33-Citation36 Cdc42, together with Cdc24, interacts with Ras1 to activate the MAPKKKK Cst20, which then triggers a subsequent phosphorylation of the MAPKKK (Ste11)-MAPKK (Hst7)-MAPK (Cek1/Cek2) cascade.Citation33-Citation36 The cascade finally activates the downstream transcription factor Cph1,Citation37 a C. albicans homolog of S. cerevisiae Ste12.Citation38 Cph1 is required for hyphal growth on solid agar, but not in liquid media.Citation37 Cst20, a C. albicans homolog of S. cerevisiae Ste20 and a kinase of p65PAK family, is essential for hyphal development and virulence.Citation28,Citation29 Although the function of C. albicans Ste11 in mating and filamentous growth has not been investigated, the kinase plays a critical role in pheromone induced biofilm formation.Citation39 Deletion of HST7, encoding a C. albicans homolog of S. serevisiae Ste7, results in defects in hyphal development. Overexpression of HST7 in the cst20Δ/Δ mutant can partially rescue its hyphal growth defect, suggesting Hst7 is downstream of Cst20.Citation28,Citation29 The kinases Cek1 and Cek2 are homologous to S. cerevisiae Kss1 and Fus3, respectively.Citation30 The mutants of hst7Δ/Δ and cph1Δ/Δ, and the cek1Δ/Δ cek2Δ/Δ double mutant are completely defective in mating, while deletion of CST20, CEK1, and CEK2 genes only leads to partial defect.Citation30,Citation31
cAMP/PKA signaling
The cAMP/PKA pathway is highly conserved in eukaryotes. In C. albicans, the cAMP/PKA pathway plays a critical role in morphogenesis.Citation40 CYR1 (also named as CDC35), encoding the only adenylyl cyclase in C. albicans, is required for hyphal development and virulence, although it is not essential for the basal level of growth.Citation41 Deletion of CYR1 has a global impact on gene expression and results in many alterations in response to stresses and environmental cues.Citation42 Cyr1 regulates serum and peptidoglycans induced filamentous growth in C. albicans.Citation11,Citation41 Recently, Huang et al. have reported that Cyr1 plays a major role in GlcNAc induced white to opaque switching, but is not essential for CO2 induction.Citation43,Citation44 The upstream activator of Cyr1 is the small GTPase Ras1, which transduces the extracellular signals to Cyr1 and stimulates cAMP production.Citation40 cAMP binds to the protein kinase A (PKA) regulatory subunits and causes their dissociation, which then activates catalytic subunits to phosphorylate downstream transcription factors. There are two catalytic subunits of PKA, namely Tpk1 and Tpk2, in C. albicans.Citation45,Citation46 The two isoforms play distinct and redundant roles in C. albicans. Inactivation of TPK1 gene causes defects in hyphal develpment on solid inducing media, but not in liquid media. In contrast, deletion of TPK2 only partially impairs hyphal growth on solid media, but has a remarkable effect in liquid media. Neither TPK1 nor TPK2 is essential for cell growth, whereas the tpk1Δ/Δ tpk2Δ/Δ double mutant is inviable.Citation45,Citation46 The catalytic subunits Tpk1 and Tpk2 are regulated by the PKA regulatory subunit Bcy1.Citation46,Citation47 In the absence of cAMP, two Bcy1 subunits bind to two catalytic subunits and inhibit PKA activity by forming an inactive heterotetrameric complex with Tpk1 or Tpk2. Null mutant of BCY1 is inviable, while it can be deleted in a tpk2/tpk2 mutant background strain.Citation47 In the presence of cAMP, binding of cAMP to Bcy1 leads to its dissociation from the complex and releases the catalytic subunits as active forms.Citation47 There are two genes, PDE1 and PDE2, encoding the low and high affinity phosphodiesterases, respectively, that keep the intracellular cAMP levels in check in C. albicans.Citation48-Citation50 Pde2 is activated in response to intracellular acidification and downregulation of cAMP signaling induced by glucose addition. Pde2 plays a major role in regulation of the intracellular levels of cAMP. Deletion of PDE2 gene results in elevated cAMP levels and constitutive activation of the cAMP pathway. Pde2 is required for normal hyphal development, but not for pseudohyphal growth. Deletion of PDE2 actually causes hyperfilamentous growth and reduced virulence.Citation48-Citation51
Efg1 and Flo8: downstream regulators of cAMP/PKA signaling
The cAMP/PKA pathway has been demonstrated to be mediated by the conserved APSES class protein Efg1 in C. albicans.Citation52,Citation53 The potential PKA phosphorylation site, threonine-206 (T206) within the conserved APSES domain of Efg1, is important for hyphal formation. Alanine substitution of T206 leads to hyphal development defect both in liquid and solid media, while glutamic acid substitution of T206 results in hyperfilamentation.Citation53 The cells of the efg1Δ/Δ mutant can only slightly elongate in response to some stimuli.Citation52 The double mutant of efg1Δ/Δ cph1Δ/Δ, representing inactivation of both the cAMP/PKA signal and the MAPK pathway, is completely locked in yeast phase under all conditions investigated.Citation52 Recently, an in vivo genome-wide ChIP-chip and in vitro footprint analysis indicates that Efg1 recognizes a sequence motif TATGCATA (EGR-box). The promoters of transcriptional regulators of hyphal growth including EFG1 itself, TCC1, CZF1, TEC1, DEF1 and NRG1 contains EGR- and/or EGR-like boxes. Further analysis demonstrates different binding specificities of Efg1 in yeast growth and in hyphal induction.Citation54
Another transcription factor downstream of cAMP/PKA pathway is the LisH domain containing protein Flo8.Citation55 C. albicans Flo8 was first identified to be essential for filamentous growth by functional complementation of an S. cerevisiae flo8Δ mutant, which is defective in invasive growth.Citation56 The C. albicans flo8Δ/Δ showed remarkably reduced virulence possibly because of its filamentous development defect. By physically interacting with Efg1, Flo8 controls expression of a subset of Efg1 regulated genes such as the hyphal growth regulator HGC1, a gene encoding a hyphal-specific G1 cyclin-related protein.Citation55,Citation57 Interestingly, the null mutants of FLO8, EFG1 and CDC35 display increased filamentation under microaerophilic conditions, suggesting that the cAMP/PKA pathway plays both positive and negative roles in regulation of morphogenesis.Citation55
pH signaling
C. albicans colonizes different niches with distinct ambient pH. For example, the normal pH of the vaginal tract is 4.5, while the pH of human blood is about 7.0. The ability of C. albicans to respond to pH changes is critical for successful colonization and infections. In C. albicans, the transcription factor Rim101 (initially named as Prr2) is the key regulator of the pH response pathway.Citation58-Citation60C. albicans Rim101 is homologous to pacC, encoding a zinc-finger transcription factor that regulates pH-dependent gene expression in the model fungus Aspergillus nidulans.Citation61,Citation62 Although deletion of RIM101 in C. albicans has no obvious effect on the cell growth at acidic or alkaline pH, the mutant shows filamentous growth defects in a number of media.Citation59,Citation60 Expression of RIM101 is pH-dependent and also controlled by Rim8 (also named as Prr1), a homolog of A. nidulans palF that regulates pacC.Citation60 The rim101Δ/Δ mutant lost the ability of controlling the expression of alkaline and acid induced genes at alkaline pH. In vivo experiments suggest that the Rim101 pathway is required for pathogenesis. The rim101Δ/Δ and rim8Δ/Δ mutants show significantly reduced virulence in a systemic infection model.Citation58
Another two pH-regulated genes are PHR1 and PHR2,encoding two cell surface glycosidases required for proper cross-linking of β-1,3- and β-1,6-glucans.Citation63,Citation64 PHR1 is expressed at pH 5.5 or higher, while PHR2 is expressed at an ambient pH below 5.5. PHR1 is upregulated in hyphal cells.Citation65 Deletion of PHR1 in C. albicans leads to growth and filamentous development defects at neutral to alkaline pHs. Conversely, deletion of PHR2 compromises filamentous growth at acidic pH.Citation63,Citation66 Consistently, the virulence phenotypes of the phr1Δ/Δ and phr2Δ/Δ mutants parallel the pH dependence of their in vitro phenotypes. As mentioned above, the systemic pH is neutral, while the vaginal pH is acidic. The phr1Δ/Δ mutant is defective in a mouse model of systemic infection but not in a rat vaginal infection, while the virulence phenotype of the phr2Δ/Δ mutant is the inverse.Citation66 The transcription of PHR1 and PHR2 is also regulated by the components of endosomal sorting complexes (ESCRT).Citation67
Contact sensing
C. albicans contact-dependent responses, including invasive growth into the host tissues and development of biofilms on surfaces, are medically important. Mkc1 is a contact-activated MAPK with a role in the cell wall integrity pathway. C. albicans mkc1Δ/Δ mutant is defective in invasive hyphal growth and biofilm development, suggesting that the cell integrity pathway plays a role in contact sensing.Citation23 The zinc finger transcription factor Czf1 is also required for contact induced response. Deletion of CZF1 in C. albicans leads to filamentous growth defect of embedded cells. Deletion of CPH1 in a czf1Δ/Δ mutant enhances this defect.Citation21 The Rho-type G protein Rac1 is highly similar to Cdc42 in protein sequence. But they have distinct roles in filamentous growth in response to different environmental conditions. In contrast to Cdc42, Rac1 is essential for filamentation induced under embedded conditions, but not required for hyphal induction by serum, GlcNAc and spider medium.Citation68,Citation69 The similar responses to different stimuli in czf1Δ/Δ and rac1Δ/Δ mutants suggest Czf1 may function downstream of Rac1.
Osmotic sensing
The Hog1 MAPK pathway regulates not only osmotic stress but also oxidative stress induced responses in C. albicans.Citation70,Citation71 This conserved signaling cascade includes three major components: the MAPKKK Ssk2, MAPKK Pbs2 and MAPK Hog1.Citation72 Ssk2 phosphorylates and thereby activates Pbs2,Citation73 which then subsequently phosphorylates the MAPK Hog1. C. albicans Hog1 was first identified by functional complementation of the osmosenstitve phenotype in an S. cerevisiae hog1Δ mutant and proven to be involved in response to osmotic stress.Citation70 Microarray analysis shows that C. albicans Hog1 plays a global role in regulation of gene transcription in response to a variety of stresses.Citation74 Deletion of HOG1 in C. albicans results in abnormal filamentous growth as well as decreased virulence.Citation75 C. albicans Pbs2, a homolog to the S. cerevisiae MAPKK Pbs2, is required for stress regulation of Hog1p localization and activity. Similar to the hog1Δ/Δ mutant, the pbs2Δ/Δ mutant is sensitive to both osmotic and oxidative stresses.Citation73 Besides the MAPKKK Ssk2, C. albicans genome sequencing reveals the existence of the upstream components of the Hog1 cascade including Ssk1, Ypd1, Chk1 and Nik1, which are homologous to their counterparts in S. cerevisiae (CGD database, www.candidagenome.org).
Negative regulators of filamentous growth
Both filamentous and yeast cells of C. albicans have been found to be associated with tissue infections. It is more likely that one form may be better adapted than the other to survive in different host niches.Citation1 Therefore, the ability of interconversion between yeast and filamentous forms is critical for pathogenesis. As aforementioned, whereas quite a lot of effort has been put into understanding the mechanisms of yeast to filamentous conversion, only a few negative regulators have been intensively studied.
The general transcriptional repressor Tup1 controls filamentous formation in C. albicans under all conditions investigated.Citation76 It represses expression of numerous genes required for initiation and maintenance of filamentous growth. Tup1 is epistatic to the transcriptional activator Cph1 since the phenotype of the tup1Δ/Δ cph1Δ/Δ double mutant is indistinguishable from that of the tup1Δ/Δ mutant.Citation76 The zinc finger transcription factor Nrg1 is a DNA-binding protein associated with Tup1.Citation77,Citation78 Deletion of NRG1 also leads to constitutive filamentous growth under all growth conditions tested. Consistently, overexpression of NRG1 repressed the yeast to filamentous transition.Citation77,Citation78 Nrg1 is thought to act by recruiting Tup1 to the promoters of target genes. Microarray data indicates that a subset of Nrg1-regulated genes is controlled by Tup1.Citation79,Citation80 Another transcriptional repressor in C. albicans is Ssn6, which acts together with Tup1. The Tup1-Ssn6 corepressor is conserved from yeast to human. In S. cerevisiae, Ssn6 forms a co-repressor complex with Tup1 and regulates a variety of genes involved in different biological processes.Citation80 Hwang et al. found that Ssn6 controls morphological conversion as well as virulence in C. albicans.Citation81 The mutant of SSN6 displays increased filamentous growth ability in response to high temperature. Interestingly, overexpression of SSN6 leads to enhanced filamentous growth and reduced virulence. These results suggest that Ssn6 may function as a repressor as well as an activator for hyphal development.Citation81 C. albicans ssn6Δ/Δ and tup1Δ/Δ mutants demonstrate distinct morphological and invasive growth phenotypes. Transcriptional profiling indicates that hypha-specific genes, which are targeted by Tup1 and Nrg1, are not derepressed in the ssn6Δ/Δ mutant, while expression of some white-opaque switching related genes (e.g., WH11) was increased.Citation80 Therefore, Ssn6 acts independent of Tup1 at least in some biological processes.
C. albicans Rfg1 is a homolog of S. cerevisiae Rox1, a key repressor of hypoxic genes. Rfg1 controls filamentous development and virulence in C. albicans, but does not appear to be required for the regulation of hypoxic genes.Citation82,Citation83 Like Nrg1, Rfg1 is a sequence-specific DNA binding protein. In S. cerevisiae, Rox1 represses expression of hypoxic genes via recruitment of the Ssn6-Tup1 complex. Rfg1 may play a similar role in C. albicans. DNA microarray analysis demonstrated that 61 genes are induced significantly in response to exposure to serum and high temperature.Citation84 Half of these genes are repressed by the transcription factors, Rfg1, Nrg1, and Tup1. Deletion of RFG1 gene in C. albicans leads to constitutively filamentous growth, while overexpression of RFG1 does not inhibit hyphal formation either in vitro or in vivo.Citation84
Cell wall proteins
The cell wall of C. albicans contains about 60% of β-glucan and about 40% of mannoproteins and chitin.Citation85,Citation86 It provides the cell with a scaffold and protection. The upstream signal transduction pathways and transcription factors finally target on the cell wall components including cell wall proteins. Cell wall proteins play critical roles in maintaining the integrity of the cell wall and sensing the external environmental cues and, therefore, are important for adaptation to the host and pathogenesis.
Hwp1 and Ywp1
Hwp1 (hyphal wall protein 1), a mannoprotein with a C-terminal GPI anchor, is exclusively expressed on the hyphal cell surface.Citation87,Citation88 HWP1 is regulated by a variety of transcription factors including Tup1, Nrg1, Efg1 and Bcr1.Citation89,Citation90 The protein Hwp1 is a substrate for the mammalian transglutaminase (TGase) and regulates covalent attachments between germ tubes and host epithelial cells.Citation87 Ywp1 (yeast wall protein 1) of C. albicans is a GPI protein containing an N-terminal secretion signal and a central region rich in serine and threonine.Citation91 The protein Ywp1 has been found to be linked covalently to the wall matrix and to accumulate in liquid culture during stationary phase. Ywp1 is regulated by environmental pH, transcriptional activator Efg1 and Efh1.Citation92 While Ywp1 is not essential for normal growth, hyphal development and virulence, deletion of YWP1 results in increased biofilm formation and adhesiveness.Citation91
ALS family proteins
The ALS (agglutinin-like sequence) gene family of C. albicans encodes cell wall-bound adhesins that are critical for biolfilm development and tissue adherence during the process of infection.Citation93 ALS family proteins are large glycoproteins with a three-domain structure, including a conserved 5′ domain, a central region of 108 bp unit of a repeated motif and a variable 3′ domain with a serine-threonine-rich sequence. There are eight ALS genes (ALS1 to ALS7 and ALS9) in C. albicans.Citation93-Citation95 ALS8 has been proven to be the same gene as ALS3. These ALS genes are located on three different chromosomes: Chr. 6 (ALS1, ALS2, ALS4 and ALS5), Chr. 3 (ALS6 and ALS7) and Chr. R (ALS3). ALS1 was first identified in C. albicans as a cell surface protein induced in response to high temperature and CO2.Citation96 The expression of ALS1 is upregulated at neutral pH and is downregulated in ssk1/ssk1 mutants. Overexpression of ALS1 leads to extensive flocculation and aggregation. ALS1 functions downstream of cAMP/PKA pathway and is targeted by the transcription factor Efg1. Consistent with its role in adhesion, expression of ALS1 is induced in biofilm populations and overexpression of ALS1 in a bcr1/bcr1 mutant rescues its biofilm development defect.Citation90 Expression of ALS genes is also differentially regulated by culture conditions, morphological form and stage of growth.Citation97,Citation98 The molecular features and roles of ALS proteins in adherence indicate their importance in virulence. Genome sequencing data show that ALS proteins also exist in other pathogenic Candida species including C. dubliniensis and C. tropicalis.Citation99
Regulation of White-Opaque Switching
The second high-frequency phenotypic switching system in C. albicans, namely white-opaque transition, has been extensively investigated during the past decade.Citation100 White and opaque phenotypes are heritable and bistable and show different cellular and colony appearances, gene expression profiles, mating ability and virulence.Citation6,Citation7,Citation101,Citation102 Cells of each phase can maintain for many generations. White cells are relatively round and form smooth hemispherical colonies on solid media, while opaque cells are large and elongated and form flat and gray or “opaque” colonies.Citation7 The two distinct types of cells express a set of phenotype-specific genes. For example, WH11 and the long transcription form of EFG1 are specifically expressed in white cells, whereas OP4 and SAP4 are enriched in opaque cells.Citation103-Citation105 Remarkably, white cells express a fermentative profile of metabolism related genes, while opaque cells adopt an oxidative one.Citation101 White and opaque cells also differ in virulence.Citation106 White cells are more virulent than opaque cells in a mouse systemic infection model and can rapidly colonize the host kidneys. In contrast, opaque cells have poor ability to colonize the host internal organs, but are better at cutaneous infection possibly due to the opaque-specific expression of secreted aspartyl proteinase (SAP) genes.
Mating type like locus (MTL)
Hull et al. discovered that C. albicans contained a mating type like locus in its genome.Citation107 The MTLa1 and MTLα2, homologous to the S. cerevisiae MATa1 and α2, have been identified on chromosome 5 in C. albicans. The MTL locus controls not only C. albicans mating but also white-opaque switching.Citation102 The Mtla1/α2 heterozygous complex inhibits white-opaque switching via controlling the expression of the master regulator gene WOR1 (white-opaque regulator 1) ().Citation108-Citation110 This finding explained why only a minor group of clinical strains could undergo white-opaque switching. This small group of strains has been proven to be homozygous at MTL locus.Citation111 In 2002, Miller et al. found that only opaque cells undergo efficient mating.Citation102 Therefore, in order to mate, C. albicans cells first have to undergo homozygosis at the MTL and then switch from white to opaque phenotype.
The white-opaque switching regulatory gene circuitry
Although the MTL homozygous strains of C. albicans are switching-competent, switching from white to opaque phase is rare and stochastic. This fact suggests that the MTL locus is not the master regulator of this process. In 2006, three labs identified Wor1 as a master regulator of the complex switching system.Citation108-Citation110 WOR1 is exclusively expressed in opaque cells and regulates its own expression by a positive feedback loop. Deletion of WOR1 locks the C. albicans cells in white phase, while overexpression of WOR1 leads to mass conversion of white to opaque form.Citation108-Citation110 In a subsequent study, Zordan et al. identified an interlocking transcriptional circuit controlling white-opaque switching.Citation112 Wor1 occupies the central position of the gene circuit, which includes three other transcription factors, Efg1, Czf1 and Wor2 (). Wor1 binds to the promoter regions of Efg1, Czf1 and Wor2 and controls their expression. Efg1 is a negative regulator of white to opaque switching. Deletion of EFG1 almost completely locks C. albicans cells in opaque phase, while overexpression of EFG1 results in opaque to white switching.Citation92,Citation105,Citation112 Overexpression of CZF1 promotes opaque cell formation possibly because it inhibits EFG1 expression. Wor2 is required for opaque formation and maintenance of opaque phenotype. The wor2Δ/Δ mutant completely lost the white to opaque switching ability.Citation112 However, ectopic expression of WOR1 in the wor2/wor2 mutant induces opaque cell formation, suggesting that Wor1 is downstream of Wor2. Microarray and RNA-seq data indicate that more than 1,000 genes or non-coding RNAs are differentially expressed in white and opaque cells, suggesting the regulation of this transition process could be much more complex.Citation101,Citation113,Citation114 Several studies suggest that epigenetic modifications of the chromatin regulate white-opaque switching in C. albicans. Treatment of C. albicans cells with the histone deacetylase inhibitor trichostatin-A (TSA) promotes opaque cell formation. Consistently, deletion of HDA1, which encodes a deacetylase sensitive to TSA, leads to increased switching frequency from white to opaque.Citation115,Citation116 Recently, Hnisz et al. have identified eight genes encoding histone-modifying enzymes as regulators of phenotypic switching.Citation117 This study indicates that the conserved Set3/Hos2 histone deacetylase complex plays a key role in white-opaque regulation and links chromatin modification to the Wor1-Wor2-Efg1-Czf1 mediated transcriptional circuit.
Environmental cues regulate white-opaque switching
A plethora of environmental cues have been found to regulate white-opaque switching in C. albicans (). Human or mammalian mucus is the natural niche for C. albicans, where the temperature is 37°C. In vitro experiments showed that opaque cells are extremely unstable and underwent mass conversion to white phase at this high temperature.Citation6 Exposure of cells to low temperature also led to opaque to white switching, but had no obvious effect on white to opaque transition.Citation6 Given opaque is the only mating competent form and is unstable at host temperature, how can C. albicans mate in the major niche of the mammalian host? Recently, Huang et al. found that two host environmental molecules, CO2 and GlcNAc, not only promoted white to opaque switching, but also could stabilize the opaque phenotype at 37°C.Citation43,Citation44 The cAMP signal, which has been proven to be essential for CO2 induced filamentous growth in C. albicans, plays a minor role in CO2 promoted white to opaque transition.Citation43 The major pathway controlling this process remains unclear. The carbonic anhydrase Nce103 is required for the low CO2 level (1%) induced opaque cell formation, but not for the high CO2 level (5%) induction.Citation43 The high levels of CO2 in the host (4.5–30.0%) may play a critical role in C. albicans phenotypic switching and sexual reproduction.Citation17 GlcNAc, a component of bacterial cell wall and GI tract mucus, is another inducer of white to opaque switching.Citation44 In contrast to CO2, GlcNAc promotes opaque phenotype primarily via the Ras1-cAMP/PKA pathway. The mutants of RAS1 and CYR1 genes showed remarkably reduced response to GlcNAc. Activating the pathway by ectopic expression of RAS1V13, a constitutively activated form of Ras1,Citation25 resulted in hypersensitivity to GlcNAc stimulation. Consistently, deletion of the high affinity phosphodiesterase gene PDE2 had the similar effect. The activated cAMP signal finally targets on the master regulator Wor1, which contains a conserved PKA phosphorylation site ().Citation44,Citation108 Interestingly, GlcNAc and CO2 have synergistic effect on induction of opaque phenotype, suggesting that these two molecules function via distinct major pathways.Citation44 Other factors, including oxidative stress, UV light and adenine, have also been found to regulate white-opaque switching, although the underlying mechanisms remain to be investigated.Citation117-Citation119
Relationship between white-opaque switching and biofilm formation
Like S. cerevisiae MATa and α cells, C. albicans MTLa/a and α/α cells secrete a or α pheromones, respectively, and carry corresponding α-factor or a-factor receptors.Citation120-Citation123 Although only opaque cells undergo efficient mating, α-pheromone induces expression of some mating-related genes, including STE2, CEK1, CEK2 and SST2, in both opaque and white cells.Citation124 The activation of the mating regulatory pathway in white cells leads to increased cohesiveness, adhesiveness and induces biofilm development. Daniels et al. demonstrate that a minority of opaque cells signal the majority of the white cell population to form biofilms, which provide an environment facilitating opaque cell mating.Citation124 The white cell response to pheromone has been shown to be a general feature of MTL-homozygous C. albicans strains via a pheromone-based paracrine system.Citation125,Citation126 Interestingly, pheromones from related Candida species can induce C. albicans white cell response, indicating the plasticity of this signal.Citation127 Recently, the Soll group has found that the Ste11-Hst7-Cek1/2-Tec1 mediated MAPK pathway primarily controls the pheromone induced biofilm development in MTL homozygous strains, while the Ras-cAMP/PKA pathway governs the conventional biofilm formed by MTLa/α strains.Citation39 Tec1 plays a central role in biofilm development of white cells, while Cph1 controls the mating response of opaque cells.Citation128 We have recently reported that the GATA type zinc finger transcription factor Gat2 plays an important role in biofilm formation, filamentation and virulence in C. albicans. We also demonstrate that Gat2 may function downstream of Tec1.Citation129
Common and distinct mechanisms of white-opaque switching and filamentous growth regulation
Opaque cells share some common features with hyphae, including a big vacuole and cell surface antigens.Citation7 It has been suggested that opaque phenotype represents a newly evolved biological process since it is unique to C. albicans and its closely related species Candida dubliniensis.Citation130 A lot of filamentous growth regulators, such as Efg1, Efh1, Czf1, Hda1 and Tup1, also control white to opaque switching.Citation21,Citation52,Citation105,Citation112,Citation116,Citation117,Citation131 Remarkably, the conserved Ras-cAMP/PKA pathway regulates CO2 and GlcNAc induced filamentous growth as well as opaque formation in C. albicans.Citation4,Citation13,Citation43,Citation44 Other environmental cues including stresses, hypoxic conditions and UV, which play a critical role in filamentous growth induction, have also been found to regulate white-opaque switching.Citation100,Citation118,Citation119,Citation132 In some aspects, opaque cells are characterized by several unique features.Citation7 First, opaque cells bud like white yeast cells although the dynamics of actin localization follow the hypha pattern later in the budding growth.Citation7 Second, the surface of opaque cells exhibits unique pimples, which are not observed in white cells and hyphae.Citation7 Third, white-opaque switching is specifically regulated by some phase related genes,Citation101,Citation113,Citation114 including the master regulator Wor1 and the zinc finger transcription factor Wor2.Citation112 Wor1 and Wor2 are not required for filamentous growth.Citation108,Citation112 Fourthly, opaque cells switch to white quickly at high temperature (37°C) in vitro, while the high temperature facilitates filamentous growth.Citation6 Finally, the white to opaque transition is a mating prerequisite in C. albicans.Citation102 Only a minor part of natural strains, which are homozygous at the MTL locus, can undergo white to opaque switching.Citation111 Therefore, common and distinct mechanisms are involved in the regulation of the two phenotypic transition systems.
Conclusion
High-frequency phenotypic transition is a defining feature of the pathogen fungus C. albicans. The ability of switching between different morphological forms in response to environmental cues is widely thought to be associated with virulence. In this review, the underlying mechanisms controlling yeast-filamentous growth transition and white-opaque switching, which represent two typical phenotypic switching systems in C. albicans, have been reviewed. The complex interplay between internal genetic elements and external environments determines the morphological fate of this organism. The genes or pathways involved in phenotypic transition are often required for virulence, indicating the important link between morphogenesis and pathogenesis in C. albicans. Interestingly, a variety of environmental inducers and genes, which regulate filamentous growth, also control white-opaque switching, suggesting that the two switching systems are evolutionarily-related. The phenotypic plasticity of C. albicans enables the organism to rapidly adapt to the changing host environments, while the biological significance of switching from white to opaque state to mate remains unclear. Given the importance of morphological changes in C. albicans, more detailed and more extensive investigations will be needed for deeper insights into understanding its pathogenicity.
Acknowledgments
The author would like to apologize to all researchers whose important work cannot be cited because of the space restrictions. The author would like to thank Drs Song Yi, Nidhi Sahni and the Huang lab members for their comments on the manuscript. Work in the Huang lab is supported by the grant of “100 Talent Program” from Chinese Academy of Sciences and the Chinese National Natural Science Foundation Grant 31170086 to G.H.
References
- Odds FC. Candida and candidosis: A Review and Bibliography. 2nd edition. W.B. Saunders Company / Bailliere Tindall, 1988.
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 2007; 20:133 - 63; http://dx.doi.org/10.1128/CMR.00029-06; PMID: 17223626
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol 2007; 61:529 - 53; http://dx.doi.org/10.1146/annurev.micro.61.080706.093341; PMID: 17506678
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev 2007; 71:348 - 76; http://dx.doi.org/10.1128/MMBR.00009-06; PMID: 17554048
- Cottier F, Mühlschlegel FA. Sensing the environment: response of Candida albicans to the X factor. FEMS Microbiol Lett 2009; 295:1 - 9; http://dx.doi.org/10.1111/j.1574-6968.2009.01564.x; PMID: 19473245
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 1987; 169:189 - 97; PMID: 3539914
- Anderson JM, Soll DR. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol 1987; 169:5579 - 88; PMID: 3316187
- Slutsky B, Buffo J, Soll DR. High-frequency switching of colony morphology in Candida albicans. Science 1985; 230:666 - 9; http://dx.doi.org/10.1126/science.3901258; PMID: 3901258
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol 2011; 9:737 - 48; http://dx.doi.org/10.1038/nrmicro2636; PMID: 21844880
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol 2004; 12:317 - 24; http://dx.doi.org/10.1016/j.tim.2004.05.008; PMID: 15223059
- Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H, et al. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 2008; 4:28 - 39; http://dx.doi.org/10.1016/j.chom.2008.05.014; PMID: 18621008
- Cassone A, Sullivan PA, Shepherd MG. N-acetyl-D-glucosamine-induced morphogenesis in Candida albicans. Microbiologica 1985; 8:85 - 99; PMID: 3883103
- Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schröppel K, Naglik JR, et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 2005; 15:2021 - 6; http://dx.doi.org/10.1016/j.cub.2005.10.040; PMID: 16303561
- Mock RC, Pollack JH, Hashimoto T. Carbon dioxide induces endotrophic germ tube formation in Candida albicans. Can J Microbiol 1990; 36:249 - 53; http://dx.doi.org/10.1139/m90-043; PMID: 2162728
- Simonetti N, Strippoli V, Cassone A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 1974; 250:344 - 6; http://dx.doi.org/10.1038/250344a0; PMID: 4605454
- Sims W. Effect of carbon dioxide on the growth and form of Candida albicans. J Med Microbiol 1986; 22:203 - 8; http://dx.doi.org/10.1099/00222615-22-3-203; PMID: 3095550
- Levitt MD, Bond JH Jr. Volume, composition, and source of intestinal gas. Gastroenterology 1970; 59:921 - 9; PMID: 5486278
- Levitt MD. Volume and composition of human intestinal gas determined by means of an intestinal washout technic. N Engl J Med 1971; 284:1394 - 8; http://dx.doi.org/10.1056/NEJM197106242842502; PMID: 5578321
- Guyton AC. Textbook of Medical Physiology. 11th edition. Saunders, 2005.
- Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida Albicans. Sabouraudia 1975; 13:148 - 53; http://dx.doi.org/10.1080/00362177585190271; PMID: 808868
- Brown DH Jr., Giusani AD, Chen X, Kumamoto CA. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol Microbiol 1999; 34:651 - 62; http://dx.doi.org/10.1046/j.1365-2958.1999.01619.x; PMID: 10564506
- Setiadi ER, Doedt T, Cottier F, Noffz C, Ernst JF. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J Mol Biol 2006; 361:399 - 411; http://dx.doi.org/10.1016/j.jmb.2006.06.040; PMID: 16854431
- Kumamoto CA. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc Natl Acad Sci U S A 2005; 102:5576 - 81; http://dx.doi.org/10.1073/pnas.0407097102; PMID: 15800048
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev 2001; 81:153 - 208; PMID: 11152757
- Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol 1999; 181:6339 - 46; PMID: 10515923
- Zhu Y, Fang HM, Wang YM, Zeng GS, Zheng XD, Wang Y. Ras1 and Ras2 play antagonistic roles in regulating cellular cAMP level, stationary-phase entry and stress response in Candida albicans. Mol Microbiol 2009; 74:862 - 75; http://dx.doi.org/10.1111/j.1365-2958.2009.06898.x; PMID: 19788542
- Leberer E, Harcus D, Dignard D, Johnson L, Ushinsky S, Thomas DY, et al. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol Microbiol 2001; 42:673 - 87; http://dx.doi.org/10.1046/j.1365-2958.2001.02672.x; PMID: 11722734
- Köhler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci U S A 1996; 93:13223 - 8; http://dx.doi.org/10.1073/pnas.93.23.13223; PMID: 8917572
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, et al. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 1996; 93:13217 - 22; http://dx.doi.org/10.1073/pnas.93.23.13217; PMID: 8917571
- Chen J, Chen J, Lane S, Liu H. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol Microbiol 2002; 46:1335 - 44; http://dx.doi.org/10.1046/j.1365-2958.2002.03249.x; PMID: 12453219
- Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol Microbiol 2002; 46:1345 - 51; http://dx.doi.org/10.1046/j.1365-2958.2002.03263.x; PMID: 12453220
- Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol 2005; 59:233 - 55; http://dx.doi.org/10.1146/annurev.micro.59.030804.121310; PMID: 15910278
- Hazan I, Liu H. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot Cell 2002; 1:856 - 64; http://dx.doi.org/10.1128/EC.1.6.856-864.2002; PMID: 12477786
- Ushinsky SC, Harcus D, Ash J, Dignard D, Marcil A, Morchhauser J, et al. CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot Cell 2002; 1:95 - 104; http://dx.doi.org/10.1128/EC.1.1.95-104.2002; PMID: 12455975
- Bassilana M, Blyth J, Arkowitz RA. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot Cell 2003; 2:9 - 18; http://dx.doi.org/10.1128/EC.2.1.9-18.2003; PMID: 12582118
- Bassilana M, Hopkins J, Arkowitz RA. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot Cell 2005; 4:588 - 603; http://dx.doi.org/10.1128/EC.4.3.588-603.2005; PMID: 15755921
- Liu H, Köhler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 1994; 266:1723 - 6; http://dx.doi.org/10.1126/science.7992058; PMID: 7992058
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 1994; 8:2974 - 85; http://dx.doi.org/10.1101/gad.8.24.2974; PMID: 8001818
- Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, et al. Alternative mating type configurations (a/α versus a/a or α/α) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol 2011; 9:e1001117; http://dx.doi.org/10.1371/journal.pbio.1001117; PMID: 21829325
- Hogan DA, Sundstrom P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol 2009; 4:1263 - 70; http://dx.doi.org/10.2217/fmb.09.106; PMID: 19995187
- Rocha CR, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN, et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 2001; 12:3631 - 43; PMID: 11694594
- Harcus D, Nantel A, Marcil A, Rigby T, Whiteway M. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol Biol Cell 2004; 15:4490 - 9; http://dx.doi.org/10.1091/mbc.E04-02-0144; PMID: 15269278
- Huang G, Srikantha T, Sahni N, Yi S, Soll DR. CO(2) regulates white-to-opaque switching in Candida albicans. Curr Biol 2009; 19:330 - 4; http://dx.doi.org/10.1016/j.cub.2009.01.018; PMID: 19200725
- Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog 2010; 6:e1000806; http://dx.doi.org/10.1371/journal.ppat.1000806; PMID: 20300604
- Bockmühl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol Microbiol 2001; 42:1243 - 57; http://dx.doi.org/10.1046/j.1365-2958.2001.02688.x; PMID: 11886556
- Cloutier M, Castilla R, Bolduc N, Zelada A, Martineau P, Bouillon M, et al. The two isoforms of the cAMP-dependent protein kinase catalytic subunit are involved in the control of dimorphism in the human fungal pathogen Candida albicans. Fungal Genet Biol 2003; 38:133 - 41; http://dx.doi.org/10.1016/S1087-1845(02)00520-0; PMID: 12553943
- Cassola A, Parrot M, Silberstein S, Magee BB, Passeron S, Giasson L, et al. Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot Cell 2004; 3:190 - 9; http://dx.doi.org/10.1128/EC.3.1.190-199.2004; PMID: 14871949
- Hoyer LL, Cieslinski LB, McLaughlin MM, Torphy TJ, Shatzman AR, Livi GP. A Candida albicans cyclic nucleotide phosphodiesterase: cloning and expression in Saccharomyces cerevisiae and biochemical characterization of the recombinant enzyme. Microbiology 1994; 140:1533 - 42; http://dx.doi.org/10.1099/13500872-140-7-1533; PMID: 8075796
- Bahn YS, Staab J, Sundstrom P. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol Microbiol 2003; 50:391 - 409; http://dx.doi.org/10.1046/j.1365-2958.2003.03692.x; PMID: 14617167
- Jung WH, Stateva LI. The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology 2003; 149:2961 - 76; http://dx.doi.org/10.1099/mic.0.26517-0; PMID: 14523128
- Jung WH, Warn P, Ragni E, Popolo L, Nunn CD, Turner MP, et al. Deletion of PDE2, the gene encoding the high-affinity cAMP phosphodiesterase, results in changes of the cell wall and membrane in Candida albicans. Yeast 2005; 22:285 - 94; http://dx.doi.org/10.1002/yea.1199; PMID: 15789349
- Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell 1997; 90:939 - 49; http://dx.doi.org/10.1016/S0092-8674(00)80358-X; PMID: 9298905
- Bockmühl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 2001; 157:1523 - 30; PMID: 11290709
- Lassak T, Schneider E, Bussmann M, Kurtz D, Manak JR, Srikantha T, et al. Target specificity of the Candida albicans Efg1 regulator. Mol Microbiol 2011; 82:602 - 18; http://dx.doi.org/10.1111/j.1365-2958.2011.07837.x; PMID: 21923768
- Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, et al. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol Biol Cell 2006; 17:295 - 307; http://dx.doi.org/10.1091/mbc.E05-06-0502; PMID: 16267276
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 1996; 144:967 - 78; PMID: 8913742
- Zheng X, Wang Y, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J 2004; 23:1845 - 56; http://dx.doi.org/10.1038/sj.emboj.7600195; PMID: 15071502
- Davis D, Edwards JE Jr., Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun 2000; 68:5953 - 9; http://dx.doi.org/10.1128/IAI.68.10.5953-5959.2000; PMID: 10992507
- Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol 2000; 20:971 - 8; http://dx.doi.org/10.1128/MCB.20.3.971-978.2000; PMID: 10629054
- El Barkani A, Kurzai O, Fonzi WA, Ramon A, Porta A, Frosch M, et al. Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol Cell Biol 2000; 20:4635 - 47; http://dx.doi.org/10.1128/MCB.20.13.4635-4647.2000; PMID: 10848590
- Espeso EA, Tilburn J, Śnchez-Pulido L, Brown CV, Valencia A, Arst HN Jr., et al. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J Mol Biol 1997; 274:466 - 80; http://dx.doi.org/10.1006/jmbi.1997.1428; PMID: 9417928
- Ramón AM, Fonzi WA. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot Cell 2003; 2:718 - 28; http://dx.doi.org/10.1128/EC.2.4.718-728.2003; PMID: 12912891
- Fonzi WA. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J Bacteriol 1999; 181:7070 - 9; PMID: 10559174
- Kurzai O, Heinz WJ, Sullivan DJ, Coleman DC, Frosch M, Mühlschlegel FA. Rapid PCR test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the pH-regulated PHR1 and PHR2 genes of C. albicans. J Clin Microbiol 1999; 37:1587 - 90; PMID: 10203530
- Melanson JE, Chisholm KA, Pinto DM. Targeted comparative proteomics by liquid chromatography/matrix-assisted laser desorption/ionization triple-quadrupole mass spectrometry. Rapid Commun Mass Spectrom 2006; 20:904 - 10; http://dx.doi.org/10.1002/rcm.2391; PMID: 16470697
- De Bernardis F, Mühlschlegel FA, Cassone A, Fonzi WA. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun 1998; 66:3317 - 25; PMID: 9632601
- Kullas AL, Li M, Davis DA. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot Cell 2004; 3:1609 - 18; http://dx.doi.org/10.1128/EC.3.6.1609-1618.2004; PMID: 15590834
- Hope H, Bogliolo S, Arkowitz RA, Bassilana M. Activation of Rac1 by the guanine nucleotide exchange factor Dck1 is required for invasive filamentous growth in the pathogen Candida albicans. Mol Biol Cell 2008; 19:3638 - 51; http://dx.doi.org/10.1091/mbc.E07-12-1272; PMID: 18579689
- Bassilana M, Arkowitz RA. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryot Cell 2006; 5:321 - 9; http://dx.doi.org/10.1128/EC.5.2.321-329.2006; PMID: 16467473
- San José C, Monge RA, Pérez-Díaz R, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J Bacteriol 1996; 178:5850 - 2; PMID: 8824643
- Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 2004; 15:4179 - 90; http://dx.doi.org/10.1091/mbc.E04-03-0181; PMID: 15229284
- Smith DA, Morgan BA, Quinn J. Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol Lett 2010; 306:1 - 8; http://dx.doi.org/10.1111/j.1574-6968.2010.01937.x; PMID: 20345377
- Arana DM, Nombela C, Alonso-Monge R, Pla J. The Pbs2 MAP kinase kinase is essential for the oxidative-stress response in the fungal pathogen Candida albicans. Microbiology 2005; 151:1033 - 49; http://dx.doi.org/10.1099/mic.0.27723-0; PMID: 15817773
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, et al. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell 2006; 17:1018 - 32; http://dx.doi.org/10.1091/mbc.E05-06-0501; PMID: 16339080
- Alonso-Monge R, Navarro-García F, Molero G, Diez-Orejas R, Gustin M, Pla J, et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol 1999; 181:3058 - 68; PMID: 10322006
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 1997; 277:105 - 9; http://dx.doi.org/10.1126/science.277.5322.105; PMID: 9204892
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. EMBO J 2001; 20:4753 - 61; http://dx.doi.org/10.1093/emboj/20.17.4753; PMID: 11532939
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J 2001; 20:4742 - 52; http://dx.doi.org/10.1093/emboj/20.17.4742; PMID: 11532938
- Murad AM, d’Enfert C, Gaillardin C, Tournu H, Tekaia F, Talibi D, et al. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol Microbiol 2001; 42:981 - 93; http://dx.doi.org/10.1046/j.1365-2958.2001.02713.x; PMID: 11737641
- García-Śnchez S, Mavor AL, Russell CL, Argimon S, Dennison P, Enjalbert B, et al. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell 2005; 16:2913 - 25; http://dx.doi.org/10.1091/mbc.E05-01-0071; PMID: 15814841
- Hwang CS, Oh JH, Huh WK, Yim HS, Kang SO. Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol Microbiol 2003; 47:1029 - 43; http://dx.doi.org/10.1046/j.1365-2958.2003.03353.x; PMID: 12581357
- Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol Cell Biol 2001; 21:2496 - 505; http://dx.doi.org/10.1128/MCB.21.7.2496-2505.2001; PMID: 11259598
- Khalaf RA, Zitomer RS. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 2001; 157:1503 - 12; PMID: 11290707
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell 2005; 16:2903 - 12; http://dx.doi.org/10.1091/mbc.E05-01-0073; PMID: 15814840
- Chaffin WL. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 2008; 72:495 - 544; http://dx.doi.org/10.1128/MMBR.00032-07; PMID: 18772287
- Iorio E, Torosantucci A, Bromuro C, Chiani P, Ferretti A, Giannini M, et al. Candida albicans cell wall comprises a branched beta-D-(1-->6)-glucan with beta-D-(1-->3)-side chains. Carbohydr Res 2008; 343:1050 - 61; http://dx.doi.org/10.1016/j.carres.2008.02.020; PMID: 18346722
- Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 1999; 283:1535 - 8; http://dx.doi.org/10.1126/science.283.5407.1535; PMID: 10066176
- Staab JF, Sundstrom P. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast 1998; 14:681 - 6; http://dx.doi.org/10.1002/(SICI)1097-0061(199805)14:7<681::AID-YEA256>3.0.CO;2-8; PMID: 9639315
- Sharkey LL, McNemar MD, Saporito-Irwin SM, Sypherd PS, Fonzi WA. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol 1999; 181:5273 - 9; PMID: 10464197
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 2005; 15:1150 - 5; http://dx.doi.org/10.1016/j.cub.2005.05.047; PMID: 15964282
- Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. Yeast wall protein 1 of Candida albicans. Microbiology 2005; 151:1631 - 44; http://dx.doi.org/10.1099/mic.0.27663-0; PMID: 15870471
- Doedt T, Krishnamurthy S, Bockmühl DP, Tebarth B, Stempel C, Russell CL, et al. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell 2004; 15:3167 - 80; http://dx.doi.org/10.1091/mbc.E03-11-0782; PMID: 15218092
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol 2001; 9:176 - 80; http://dx.doi.org/10.1016/S0966-842X(01)01984-9; PMID: 11286882
- Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family--a sticky pursuit. Med Mycol 2008; 46:1 - 15; http://dx.doi.org/10.1080/13693780701435317; PMID: 17852717
- Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet 1998; 33:451 - 9; http://dx.doi.org/10.1007/s002940050359; PMID: 9644209
- Donohue DS, Ielasi FS, Goossens KV, Willaert RG. The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans. Mol Microbiol 2011; 80:1667 - 79; http://dx.doi.org/10.1111/j.1365-2958.2011.07676.x; PMID: 21585565
- Giacometti R, Kronberg F, Biondi RM, Passeron S. Candida albicans Tpk1p and Tpk2p isoforms differentially regulate pseudohyphal development, biofilm structure, cell aggregation and adhesins expression. Yeast 2011; 28:293 - 308; http://dx.doi.org/10.1002/yea.1839; PMID: 21234881
- Sosinska GJ, de Koning LJ, de Groot PW, Manders EM, Dekker HL, Hellingwerf KJ, et al. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 2011; 157:136 - 46; http://dx.doi.org/10.1099/mic.0.044206-0; PMID: 20864472
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009; 459:657 - 62; http://dx.doi.org/10.1038/nature08064; PMID: 19465905
- Soll DR. Why does Candida albicans switch?. FEMS Yeast Res 2009; 9:973 - 89; http://dx.doi.org/10.1111/j.1567-1364.2009.00562.x; PMID: 19744246
- Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, et al. Metabolic specialization associated with phenotypic switching in Candidaalbicans. Proc Natl Acad Sci U S A 2002; 99:14907 - 12; http://dx.doi.org/10.1073/pnas.232566499; PMID: 12397174
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 2002; 110:293 - 302; http://dx.doi.org/10.1016/S0092-8674(02)00837-1; PMID: 12176317
- Morrow B, Srikantha T, Anderson J, Soll DR. Coordinate regulation of two opaque-phase-specific genes during white-opaque switching in Candida albicans. Infect Immun 1993; 61:1823 - 8; PMID: 8478072
- Srikantha T, Soll DR. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 1993; 131:53 - 60; http://dx.doi.org/10.1016/0378-1119(93)90668-S; PMID: 7916716
- Srikantha T, Tsai LK, Daniels K, Soll DR. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J Bacteriol 2000; 182:1580 - 91; http://dx.doi.org/10.1128/JB.182.6.1580-1591.2000; PMID: 10692363
- Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun 1999; 67:6652 - 62; PMID: 10569787
- Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 1999; 285:1271 - 5; http://dx.doi.org/10.1126/science.285.5431.1271; PMID: 10455055
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci U S A 2006; 103:12813 - 8; http://dx.doi.org/10.1073/pnas.0605270103; PMID: 16905649
- Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell 2006; 5:1674 - 87; http://dx.doi.org/10.1128/EC.00252-06; PMID: 16950924
- Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A 2006; 103:12807 - 12; http://dx.doi.org/10.1073/pnas.0605138103; PMID: 16899543
- Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, et al. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 2002; 162:737 - 45; PMID: 12399384
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol 2007; 5:e256; http://dx.doi.org/10.1371/journal.pbio.0050256; PMID: 17880264
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 2003; 115:389 - 99; http://dx.doi.org/10.1016/S0092-8674(03)00885-7; PMID: 14622594
- Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, et al. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet 2010; 6:e1001070; http://dx.doi.org/10.1371/journal.pgen.1001070; PMID: 20808890
- Klar AJ, Srikantha T, Soll DR. A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics 2001; 158:919 - 24; PMID: 11404352
- Srikantha T, Tsai L, Daniels K, Klar AJ, Soll DR. The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J Bacteriol 2001; 183:4614 - 25; http://dx.doi.org/10.1128/JB.183.15.4614-4625.2001; PMID: 11443097
- Hnisz D, Schwarzmüller T, Kuchler K. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol Microbiol 2009; 74:1 - 15; http://dx.doi.org/10.1111/j.1365-2958.2009.06772.x; PMID: 19555456
- Morrow B, Anderson J, Wilson J, Soll DR. Bidirectional stimulation of the white-opaque transition of Candida albicans by ultraviolet irradiation. J Gen Microbiol 1989; 135:1201 - 8; PMID: 2695598
- Alby K, Bennett RJ. Stress-induced phenotypic switching in Candida albicans. Mol Biol Cell 2009; 20:3178 - 91; http://dx.doi.org/10.1091/mbc.E09-01-0040; PMID: 19458191
- Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol 2003; 23:8189 - 201; http://dx.doi.org/10.1128/MCB.23.22.8189-8201.2003; PMID: 14585977
- Lockhart SR, Zhao R, Daniels KJ, Soll DR. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot Cell 2003; 2:847 - 55; http://dx.doi.org/10.1128/EC.2.5.847-855.2003; PMID: 14555467
- Panwar SL, Legrand M, Dignard D, Whiteway M, Magee PT. MFalpha1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot Cell 2003; 2:1350 - 60; http://dx.doi.org/10.1128/EC.2.6.1350-1360.2003; PMID: 14665468
- Dignard D, El-Naggar AL, Logue ME, Butler G, Whiteway M. Identification and characterization of MFA1, the gene encoding Candida albicans a-factor pheromone. Eukaryot Cell 2007; 6:487 - 94; http://dx.doi.org/10.1128/EC.00387-06; PMID: 17209123
- Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J 2006; 25:2240 - 52; http://dx.doi.org/10.1038/sj.emboj.7601099; PMID: 16628217
- Sahni N, Yi S, Pujol C, Soll DR. The white cell response to pheromone is a general characteristic of Candida albicans strains. Eukaryot Cell 2009; 8:251 - 6; http://dx.doi.org/10.1128/EC.00320-08; PMID: 19074600
- Yi S, Sahni N, Daniels KJ, Lu KL, Huang G, Srikantha T, et al. Self-induction of a/a or alpha/alpha biofilms in Candida albicans is a pheromone-based paracrine system requiring switching. Eukaryot Cell 2011; 10:753 - 60; http://dx.doi.org/10.1128/EC.05055-11; PMID: 21498642
- Alby K, Bennett RJ. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci U S A 2011; 108:2510 - 5; http://dx.doi.org/10.1073/pnas.1017234108; PMID: 21262815
- Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, Soll DR. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol 2010; 8:e1000363; http://dx.doi.org/10.1371/journal.pbio.1000363; PMID: 20454615
- Du H, Guan G, Xie J, Sun Y, Tong Y, Zhang L, et al. Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One 2012; 7:e29707; http://dx.doi.org/10.1371/journal.pone.0029707; PMID: 22276126
- Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, et al. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell 2004; 3:1015 - 27; http://dx.doi.org/10.1128/EC.3.4.1015-1027.2004; PMID: 15302834
- Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol 2011; 9:e1001105; http://dx.doi.org/10.1371/journal.pbio.1001105; PMID: 21811397
- Ramírez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhäuser J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog 2008; 4:e1000089; http://dx.doi.org/10.1371/journal.ppat.1000089; PMID: 18551173
- Lane S, Zhou S, Pan T, Dai Q, Liu H. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol Cell Biol 2001; 21:6418 - 28; http://dx.doi.org/10.1128/MCB.21.19.6418-6428.2001; PMID: 11533231
- Schweizer A, Rupp S, Taylor BN, Röllinghoff M, Schröppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol 2000; 38:435 - 45; http://dx.doi.org/10.1046/j.1365-2958.2000.02132.x; PMID: 11069668
- Kumar MJ, Jamaluddin MS, Natarajan K, Kaur D, Datta A. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc Natl Acad Sci U S A 2000; 97:14218 - 23; http://dx.doi.org/10.1073/pnas.250452997; PMID: 11114181
- Yamada-Okabe T, Sakamori Y, Mio T, Yamada-Okabe H. Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur J Biochem 2001; 268:2498 - 505; http://dx.doi.org/10.1046/j.1432-1327.2001.02135.x; PMID: 11298769
- Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol Biol Cell 2007; 18:965 - 75; http://dx.doi.org/10.1091/mbc.E06-10-0931; PMID: 17192409
- Stichternoth C, Ernst JF. Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans. Appl Environ Microbiol 2009; 75:3663 - 72; http://dx.doi.org/10.1128/AEM.00098-09; PMID: 19346360
- Stichternoth C, Fraund A, Setiadi E, Giasson L, Vecchiarelli A, Ernst JF. Sch9 kinase integrates hypoxia and CO2 sensing to suppress hyphal morphogenesis in Candida albicans. Eukaryot Cell 2011; 10:502 - 11; http://dx.doi.org/10.1128/EC.00289-10; PMID: 21335533
- Bastidas RJ, Heitman J, Cardenas ME. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog 2009; 5:e1000294; http://dx.doi.org/10.1371/journal.ppat.1000294; PMID: 19197361
- Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 2008; 67:47 - 62; http://dx.doi.org/10.1111/j.1365-2958.2007.06013.x; PMID: 18078440