Adherence of Plasmodium falciparum-infected erythrocytes to the endothelium is associated with morbidity and mortality from malaria in humans. The parasite expresses a variant protein on the surface of infected erythrocytes called PfEMP1 responsible for adherence. Monoallelic transcription and expression of one of 60 var genes encoding this family in each parasite genome, and the ability to switch expression between them, provides a mechanism of antigenic variation. In a recent study, we reported the identification of a histone methyltransferase, called PfSET10, which localizes to the perinuclear active var gene transcription zone. PfSET10 is required for var gene repression and maintenance of the active var gene in the poised state during division for activation in daughter parasites. The identification of the PfSET10 molecule is the first step toward understanding how the expression of PfEMP1 protein is regulated and produced by the parasite. With improved insight into the systems, including the molecules that are involved in controlling antigenic variation, this opens the possibility of targeted treatments that would be more effective in preventing malaria infection.
The malaria parasite Plasmodium falciparum infects humans and is responsible for the majority of infection and fatal outcomes occurring in Sub-Saharan Africa. The major virulence factor is the highly polymorphic P. falciparum Erythrocyte Membrane protein 1 (PfEMP1), a protein expressed by the parasite on the surface of the red blood cell, where it mediates adherence of P. falciparum-infected erythrocytes to receptors on endothelial cells (Baruch et al., Cell 1995; Su et al., Cell 1995). Sequestration of infected erythrocytes to receptors on the endothelial lining of blood vessels, erythrocytes and platelets prevent clearance through the spleen (Barnwell, Exp Parasitol 1989; Berendt et al., Nature 1989; Cooke and Coppel, Parasitol Today 1995; Ockenhouse et al., J Exp Med 1992; Rogerson et al., J Exp Med 1995; Wahlgren et al., Parasitol Today 1994). Moreover, this can lead to obstruction of the microvasculature in organs, playing an important role in malaria pathogenesis (Leech et al., J Exp Med 1984).
Each parasite genome contains approximately 60 var genes that encode the PfEMP1 family (Gardner et al., Nature 2002). They are located at the subtelomeric or central regions of chromosomes and their transcription is mutually exclusive; with one being transcribed at any given time and the remaining copies are silenced. Antigenic variation is maintained through switching of expression as a result of activation of a different var gene, resulting in the production of variant PfEMP1 molecules with alternating receptor-binding properties (Baruch et al., Cell 1995; Smith et al., Cell 1995; Su et al., Cell 1995).
Much of our current knowledge on var gene regulation is based on the study of parasites selected for the expression of one PfEMP1 molecule transcribed from a single var locus. The analysis of one selected var locus in its active and silenced state has provided insights into histone modification of chromatin and the discovery of chromatin binding and/or modifying molecules involved in var gene regulation. Despite some important progress our current understanding on the molecular players and events determining var gene activation, poising, silencing and switching is limited (Duffy et al., Cell Microbiol 2012).
In an attempt to discover novel regulators for var gene expression, we localized a number of putative epigenetic factors to the P. falciparum nucleus (Volz et al., IJP 2010). The Plasmodium-specific SET-domain protein PFL1010c, now called PfSET10, showed an intriguing localization exclusively to one spot within the nuclear periphery. Studies in yeast previously showed the association of functional compartments within the nuclear periphery with active genes (Akhtar and Gasser, Nat Rev Genet 2007) and further analysis into the PfSET10 localization revealed its association with euchromatin, characterized by the presence of histone marks H3K4me1, me2, me3 and H3K9ac. Var gene expression is initiated in the early ring stage parasite 4 h post invasion and involves var locus translocation to an expression site (Duraisingh et al., Cell 2005; Freitas-Junior et al., Cell 2005; Ralph et al., Proc Natl Acad Sci USA 2005; Dzikowski et al., EMBO Rep 2007), which consists of a defined zone of relaxed euchromatin at the nuclear periphery free of the heterochromatic histone mark H3K9me3 and heterochromatin protein 1. With the discovery of PfSET10 we were able to associate the first epigenetic regulator to the var gene expression site ().
Figure 1. A proposed model of PfSET10 regulation of the active var gene in a perinuclear compartment in Plasmodium falciparum (see text for details).
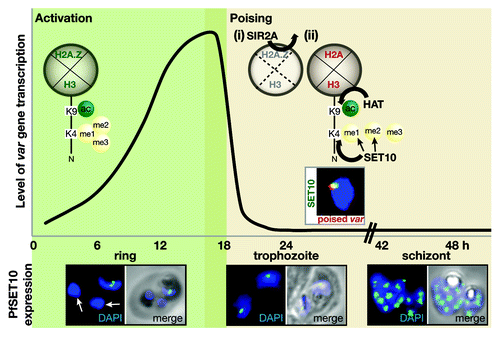
During transcription, the var promoter is enriched in histone marks H3K9ac and H3K4me3 as well as the histone variant H2A.Z (Lopez-Rubio et al., Mol Microbiol 2007; Bártfai et al., PLoS Pathog 2010; Petter et al., PLoS Pathog 2011). As the parasite enters the trophozoite stage, var gene transcription is deactivated. This step is consistent with the depletion of the histone variant H2A.Z by the histone deacetylase PfSir2A at the active var promoter (Petter et al., PLoS Pathog 2011). After parasite DNA replication and differentiation, canonical histones such as H2A and H3 are deposited, which can be already acetylated prior to nucleosome assembly (Corpet and Almouzni, Trends Cell Biol 2009), while lacking methylation marks (Loyola et al., Mol Cell 2006). Thus, the incorporation of fresh nucleosomes at the var gene promoter consisting of a new set of core histones provides the opportunity of silencing of the active var gene and switching expression to another family member. Var gene silencing is concurrent with the methylation of the histone H3K9 (Lopez-Rubio et al., Mol Microbiol 2007), subsequent recruitment of Heterochromatin Protein 1 to the var locus (Flueck et al., PLoS Pathog 2009; Pérez-Toledo et al., Nucleic Acid Res 2009) and translocation out of the ‘expression site’ (Ralph et al., Proc Natl Acad Sci USA 2005). In contrast, the poised var gene continues to be enriched in its promoter for histone marks H3K4me2, H3K4me3 and H3K9ac during division to ensure transcription in daughter cells and further remains within the “expression site” marked by the presence of PfSET10. Based on our recent findings, our current model envisages that PfSET10 is responsible for setting the H3K4me2 and H3K4me3 mark at the active var gene to maintain it in its poised state. PfSET10's specific localization to a small and defined zone within the nuclear periphery implies the presence of a very specialized and controlled mechanism of PfSET10 recruitment. Initial clues were provided by the analysis of the histone-binding properties of the PfSET10 PHD finger domain. When presented with differently modified and unmodified histone H3 peptides in an in vitro set-up, the PfSET10 PHD finger displayed a distinct affinity to the un-methylated and mono-methylated form of histone H3K4. These findings imply that one mode of PfSET10 recruitment lies in the PHD finger binding of lysine 4 of a histone H3, which has been freshly in-cooperated into a new nucleosome. After setting the methyl-marks on lysine 4 of histone H3, PfSET10 withdraws from its site of action, possibly through the rapid decrease of its PHD finger affinity to the methylated forms of histone H3K4. A further potential PfSET10 recruitment factor, which is deposited into a newly formed nucleosome, is histone H2A. Histone H2A replaces H2A.Z after var gene transcription is turned off and is a characteristic feature of poised or silenced var gene promoters (Petter et al., PLoS Pathog 2011). While PfSET10 recruitment to H2A mediated via its PHD finger appears likely, this hypothesis requires further attention. It is tempting to speculate that a combination of specific histone H2A, H3 and H4 core residues are required for PfSET10 binding as this has recently been shown to be a prerequisite for histone methyltransferase recruitment (Du and Briggs, J Biol Chem 2010). While a specifically formed nucleosomal surface may serve as a PfSET10 recognition site, DNA-specific binding factors may also contribute significantly to its recruitment. Interestingly, we detected PfActin-1 as a PfSET10 interacting factor and as such a component of the var gene expression site. PfActin-1 has recently been shown to be directly involved in var gene positioning and a spatial chromosome organization (Zhang et al., Cell Host Microbe 2011). Based on these observations, PfActin-1’s role is similar and appears to be conserved to what has been observed in other eukaryotes. Whether PfActin-1 recruits PfSET10 directly or if it is responsible for positioning the active var gene into the transcription site and by this forms an interaction with PfSET10 remains to be elucidated. Besides PfActin-1, our protein-interaction analysis identified further potential PfSET10 interacting partners, such as putative RNA and DNA/RNA binding proteins. While these interesting candidates for PfSET10 interaction still await functional characterization, it is an exciting thought, that with the discovery of PfSET10, we have found a stepping stone, from which the further molecular and functional dissection of the var gene expression is feasible.
However, the physically small size of the var gene expression site and the concentrated but scarce PfSET10 protein amount per nucleus clearly present technical challenges in terms of molecular analyses. Var gene expression is a highly dynamic process and it may be difficult to catch the players in the act. Several attempts to chromatin-immunoprecipitate PfSET10 failed. With no enrichment observed at the active var gene (ChIP-PCR), or at any other parts of the genome (ChIP-seq) points once more to the generally temporal and spatial association of histone modifying enzymes with their target sites.
In recent years, histone modifying enzymes have been intensively studied, since their potential as drug targets for a range of diseases has been recognized. Large chemical libraries have been screened to identify specific histone methyltransferase inhibitors. To date, two selective inhibitors of mammalian histone lysine methyltransferase SU(VAR)3–9 and G9a, respectively, have been discovered (Greiner et al., Nat Chem Biol 2005; Kubicek et al., Mol Cell 2007). With the emergence of drug resistance in P. falciparum, drug companies have only recently started to contribute chemical libraries which has led to detection of two new promising anti-malarial compounds, OZ439 and NITD609 (Charman et al., Proc Natl Acad Sci USA 2011; Rottmann et al., Science 2010). PfSET10 is a conserved gene within the Apicomplexan lineage and based on its sequence and structural information its SET domain does not show any similarity to other known histone lysine methyltransferases. This feature places PfSET10 as an interesting candidate for drug screening since an inhibitor would probably display high specificity for PfSET10 binding with a potential to additionally target PfSET10 homologs of other Plasmodium species. Further, PfSET10 is essential to P. falciparum survival. Several attempts to disrupt the pfset10 locus or destabilize PfSET10 protein expression were unsuccessful. Interestingly, this observation also implies that the three other P. falciparum SET-domain proteins, PfSET1, PfSET4 and PfSET6, which have been predicted to target histone H3 lysine 4 (Cui et al., Int J Parasitol 2008) are unlikely to be functionally redundant with that of PfSET10. This hypothesis is further supported by the apparently different localization of PfSET6 and PfSET4 within the nucleus (Volz et al., Int J Parasitol 2010). Nevertheless, additional enzymes or co-factors may be required for proper PfSET10 activity.
The essentiality of PfSET10 to the parasite indicates that it may not merely be responsible for var gene regulation per se, but instead harbors a transcription site probably utilized by additional genes. The removal of PfSET10 from its transcription site may likely compromise its functionality and subsequently result in altered or abolished expression of potentially essential proteins. Also, a PfSET10 deficiency and resulting lack of preservation of the local euchromatic environment, may in turn affect overall chromosome organization and function.
Targeting unique components of the var gene expression site and by this interfering with the parasite’s virulence and antigenic variation program would ideally leave the parasite with a compromised ability to evade protective mechanisms of the host, enabling the immune system to take charge. We have identified a histone H3 lysine 4 methyltransferase, PfSET10, which is the first protein shown to regulate variant PfEMP1 expression. It is unique and essential to the parasite, thus representing a potential target for intervention and thereby control of this devastating disease in humans.