Fibronectin (FN) is a large and essential glycoprotein present in the blood plasma, body fluids and extracellular matrix of vertebrates.Citation1 It is a disulfide-linked dimer of nearly identical subunits, each with 12 Fn1, two Fn2 and 15–17 Fn3 modules. The Fn1 module folds into a small globule comprising an anti-parallel AB β-sheet that forms a sandwich with a more extensive anti-parallel CDE β-sheet; the sheets enclose a small hydrophobic core and the structure is further stabilized by two disulfide bridges linking the A and D strands and the D and E strands.Citation2 Neither the Fn1 module nor a FN-like protein has been identified outside of chordates, and the conceptual FN-like protein that is found in Ciona, a primitive chordate, is quite different from vertebrate FN.Citation3 FN is the only protein with tandem Fn1 modules. FN and its tandem FN1 modules, therefore, are a vertebrate invention with which bacteria have had 500 million years to adapt. This time has not been wasted. A recent review by Williams et al. introduced the reader to over 100 different fibronectin-binding proteins from over 45 Gram-positive or Gram-negative species.Citation4
These proteins are displayed on surfaces of bacteria, and various ones interact with different sites on fibronectin.Citation4 A favorite site involves the tandem Fn1 modules that are grouped at the N-termini of the fibronectin subunit. As shown in , an otherwise intrinsically disordered stretch of the bacterial protein converts to a fourth β-strand on top of the CDE sheet. Multiple Fn1 modules are thus ligated, forming a large and highly elongated interface. The binding energy, dominated by favorable enthalpy overcoming unfavorable entropy, is distributed throughout the interface.Citation5 This novel form of protein-protein interaction, called the tandem β-zipper, exploits the promiscuous display of edges of CDE β-sheets of multiple tandem Fn1 modules.
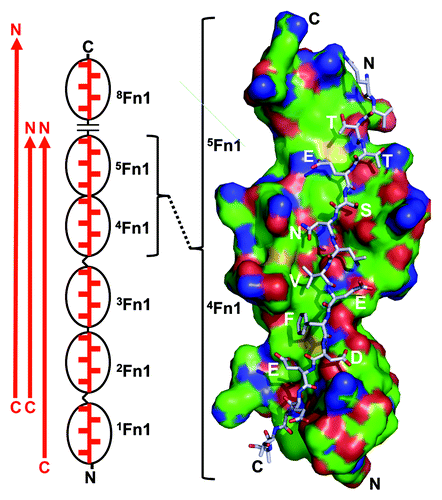
Figure 1. Tandem β-zipper formation of segments of bacterial surface proteins with N-terminal Fn1 modules of fibronectin. The segments, which by themselves are intrinsically disordered and elongated (red arrow), bind to multiple Fn1 modules (black ovals). Binding is driven by backbone hydrogen bonds between the segment and E-strands of the Fn1 modules and favorable interactions of segment side chains with the surface of the modules. Such interactions are illustrated on the right with a space-filling model of 4,5Fn1 interacting with a stick model of a segment of S. aureus FnBPA (based on crystal structure PDB 2RL0_BC12). Note that stick model zigzags through a broad binding groove with alternate side chains (indicated in 1-letter code) extending perpendicularly to either side. Segments have been identified that interact with 1-5Fn1, 2-5Fn1, or 2-5Fn1 and 8Fn1 (left). When the interacting side chains are included, the arrangement does indeed look like a zipper (left). N- and C-termini are labeled to emphasize the anti-parallel nature of the interaction.
Functions of bacteria-fibronectin interactions remain murky. The interaction affords bacteria the means to adhere to extracellular matrix and access to fibronectin-binding cell surface proteins that mediate cellular uptake of fibronectin. Such events may allow bacteria to seed tissues and evade host defense mechanisms by hiding inside cells. Alternatively, the host may use fibronectin to control bacteria. Mortality of mice after intra-peritoneal injection with Streptococcus pyogenes bearing or lacking the major F1 fibronectin-binding protein suggests that the interaction favors the host.Citation6 Protein F1-expressing bacteria were less virulent in the normal mice than bacteria not expressing protein F1, and there was a trend to protein F1-expressing bacteria being less virulent in normal mice than in mice lacking plasma fibronectin. The balance, however, may be different in other infections. Isolates of Staphylococcus aureus recovered from patients with infected cardiac devices have been found to harbor polymorphisms in fibronectin-binding protein A that are associated with stronger and more resilient bacterial binding to fibronectin, presumably by enhancing β-zipper formation.Citation7 This finding suggests that stronger binding to fibronectins selects for S. aureus that can form infectious biofilms on implanted devices.
Can interactions of bacteria with host components be targeted therapeutically? The paper by Krachler et al. appearing in this issueCitation8 is the latest in a series that indicates such a strategy may be viable. The study is based on multivalent adhesion molecule 7 (MAM7), which the authors characterized in 2011 as promoting adhesion of a range of Gram-negative pathogens to vertebrate cells by virtue of interactions with fibronectin and phosphatidic acid.Citation9,Citation10 The N-terminal 30-kDa fragment of fibronectin that comprises 1-5Fn1 was found to be sufficient for MAM7 binding.Citation10 Whether MAM7 interacts with the 30-kDa by β-zipper formation has not been determined. Remarkably, MAM7 does not appear in the wide-ranging 2011 review of bacterial fibronectin-binding proteins written by Henderson et al.Citation4
MAM7 was found to be necessary for virulence of Vibrio parahaemolyticus in a nematode infection model.Citation9 Further, efficient killing of cultured mammalian cells by V. parahaemolyticus, V. cholerae, Yersenia pseudotuberculosis or enteropathogenic Escherichia coli was less when non-pathogenic E. coli expressing surface MAM7 or polymer beads coated with recombinant MAM7 were included in the incubations.Citation9 The concept of countering Gram-negative pathogens with particles armed with their own outer membrane adhesion receptor was further developed in a Virulence article earlier this year that was entitled “Turnabout is fair play….”Citation11
Krachler et al. now have tested the concept with Acinetobacter baumannii-calcoaceticus, Pseudomonas aeruginosa and extended spectrum β-lactamase-producing E. coli and Klebsiella pneumoniae cultured from combat wounds. Five isolates of each of the pathogens were tested for cytotoxicity toward cultured human HeLa or mouse 3T3 cells in the absence or presence of MAM7-expressing non-pathogenic E. coli or MAM7-coated beads. All isolates were demonstrated to have multi-drug resistance. In the absence of MAM7-bearing particles, bacterial attachment in general correlated with cytotoxicity. Impressively, the particles decreased cytotoxicity of 19 of the 20 isolates. Cytotoxicity was quantified by release of lactate dehydrogenase and confirmed by fluorescence microscopy. In cell layers incubated with MAM7-coated beads, the beads associated with cell surfaces, presumably binding to sites on fibronectin that would otherwise be targeted by MAM7-expressing bacteria.
The authors suggest several directions for future research but tout the most important conclusion of the paper as being the demonstration that the culture assays may serve as a useful predictor of the success of MAM7-based inhibitors and other potential inhibitors that are based on the “turnabout” strategy. This surely is true. Bacterial fibronectin-binding proteins, like MAM7, usually interact with other extracellular molecules as well as with fibronectin.Citation4 Will MAM7-based inhibitors attenuate virulence mediated by other bacterial fibronectin-binding proteins? Can the strategy be used with bacterial proteins that recognize other host extracellular components? The potential targets in such experiments are immense. We need facile assays to sort through the many permutations and make up for our 500 million year handicap.
References
- Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci 2002; 115:3861 - 3; http://dx.doi.org/10.1242/jcs.00059; PMID: 12244123
- Bingham RJ, Potts JR. Fibronectin structure: a new piece of the puzzle emerges. Structure 2010; 18:660 - 1; PMID: 20541501
- Tucker RP, Chiquet-Ehrismann R. Evidence for the evolution of tenascin and fibronectin early in the chordate lineage. Int J Biochem Cell Biol 2009; 41:424 - 34; http://dx.doi.org/10.1016/j.biocel.2008.08.003; PMID: 18761101
- Henderson B, Nair S, Pallas J, Williams MA. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol Rev 2011; 35:147 - 200; http://dx.doi.org/10.1111/j.1574-6976.2010.00243.x; PMID: 20695902
- Norris NC, Bingham RJ, Harris G, Speakman A, Jones RP, Leech A, et al. Structural and functional analysis of the tandem β-zipper interaction of a Streptococcal protein with human fibronectin. J Biol Chem 2011; 286:38311 - 20; http://dx.doi.org/10.1074/jbc.M111.276592; PMID: 21840989
- Nyberg P, Sakai T, Cho KH, Caparon MG, Fässler R, Björck L. Interactions with fibronectin attenuate the virulence of Streptococcus pyogenes. EMBO J 2004; 23:2166 - 74; http://dx.doi.org/10.1038/sj.emboj.7600214; PMID: 15103329
- Lower SK, Lamlertthon S, Casillas-Ituarte NN, Lins RD, Yongsunthon R, Taylor ES, et al. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci U S A 2011; 108:18372 - 7; http://dx.doi.org/10.1073/pnas.1109071108; PMID: 22025727
- Krachler AM, Mende K, Murray C, Orth K. In vitro characterization of Multivalent Adhesion Molecule 7-based inhibition of multidrug-resistant bacteria isolated from wounded military personnel. Virulence 2012; 3:390 - 99; http://dx.doi.org/10.4161/viru.20816; PMID: 22722243
- Krachler AM, Ham H, Orth K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc Natl Acad Sci U S A 2011; 108:11614 - 9; http://dx.doi.org/10.1073/pnas.1102360108; PMID: 21709226
- Krachler AM, Orth K. Functional characterization of the interaction between bacterial adhesin multivalent adhesion molecule 7 (MAM7) protein and its host cell ligands. J Biol Chem 2011; 286:38939 - 47; http://dx.doi.org/10.1074/jbc.M111.291377; PMID: 21937438
- Krachler AM, Ham H, Orth K. Turnabout is fair play: use of the bacterial Multivalent Adhesion Molecule 7 as an antimicrobial agent. Virulence 2012; 3:68 - 71; http://dx.doi.org/10.4161/viru.3.1.18172; PMID: 22086133
- Bingham RJ, Rudiño-Piñera E, Meenan NA, Schwarz-Linek U, Turkenburg JP, Höök M, et al. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc Natl Acad Sci U S A 2008; 105:12254 - 8; http://dx.doi.org/10.1073/pnas.0803556105; PMID: 18713862