Abstract
Streptococcus pyogenes (GAS) is a human pathogen that causes multiple infections worldwide. The pathogenic properties of GAS strains are often linked to the production of virulence factors such as toxins, proteases or DNases. Detection of virulence factors produced by GAS strains can be used to either determine pathogenic potential of the strain or as a rapid screening and typing method. We recently developed a method to detect simultaneously 20 GAS virulence factors (spd3, sdc, sdaB, sdaD, speB, spyCEP, scpA, mac, sic, speL, K, M, C, I, A, H, G, J, smeZ and ssa) in four low volume multiplex PCR reactions (Borek et al., 2011) and below we present a detailed protocol describing the method.
Introduction
Streptococcus pyogenes (GAS) is a human pathogen that causes over 600 million of infections worldwide that result in over 500,000 deaths a year.Citation1 GAS causes mild infections of skin and mucosal surfaces, but is also able to cause severe, life threatening invasive diseases such as streptococcal toxic shock syndrome (STSS) or necrotizing fasciitis (NF).Citation2
Severity of GAS infections depends on multiple bacterial and host factors. The pathogenic properties of GAS strains are often linked to the production of virulence factors such as toxins, proteases or DNases and toxins present in the particular GAS strain can be the predictor of its invasiveness.Citation3,Citation4
Virulence factors are not equally distributed among S. pyogenes strains. Some of them are chromosomally encoded; others are related to the presence of mobile genetic elements. Profile of the virulence factors encoded by the particular strain is part of strain characteristics and their presence/absence can be used as diagnostic method and simple tool in clinical diagnosis.
So far, determination how S. pyogenes strains/isolates are related to each other has been rather expensive and time consuming task. To determine whether strains are related and what their pathogenic properties are, researchers and epidemiologists use usually restriction analysis of large chromosomal fragments—PFGE. PFGE is time-consuming, requires well trained personnel and the results are difficult to compare between laboratories. Other methods such as emm typing or MLST are based on the sequencing of one to several genes. Although, sequencing based methods are relatively easy to perform and easily comparable, cost of sequencing reactions can be too high for routine use. Because of these multiple disadvantages, new typing methods and schemes usually based on simple PCR reactions are proposed. A few years ago, multiplex PCR system that detects 11 superantigens has been described. The method is using SYBRGreen detection of small PCR products and requires real time PCR instrument with module for melting curve analysis.Citation5
We recently developed a method that expands methods like those mentioned above, the set of four low volume multiplex PCR reactions detect simultaneously 20 GAS virulence factors, not only superantigens, (spd3, sdc, sdaB, sdaD, speB, spyCEP, scpA, mac, sic, speL, K, M, C, I, A, H, G, J, smeZ and ssa).Citation6 The method has high resolution with Simpson’s index of diversity 0.943 (0.936–0.951) and can be used for both typing and detection of virulence factors.
Reagents
(1) Lysozyme stock 20 mg/ml (Sigma-Aldrich, L6876)
(2) Mutanolysin stock 10 U/μl (Sigma-Aldrich, M9901)
(3) RNase stock 10 mg/ml (Sigma-Aldrich, R5503)
(4) 1 mM stock of dNTP prepared from 100 mM stock solutions of individual dNTPs (Sigma-Aldrich, DNTP100)
(5) Taq polymerase (Fermentas, EP0402)
(6) 10× Taq buffer with (NH4)2SO4 (Fermentas, B33)
(7) 25 mM MgCl2 stock solution (Fermentas, R0971)
(8) Starters (see ; Genomed, www.genomed.pl)
Table 1. Starters for the detection of GAS virulence factors
(9) 1.5% SeaKemLE agarose (Lonza, 90004) in 1× TBE buffer
(10) 1× TBE electrophoresis buffer
Equipment
(1) Veriti or 9700 PCR by Life Technologies (formerly Applied Biosystems). Other PCR instruments were not tested.
(2) Gel electrophoresis Sub-Cell tank Model 192 (BioRad, 170-4509) with 26 well combs (BioRad, 170-4526)
Setup
Template
As a template for PCR reaction we use chromosomal DNA isolated using commercially available kits. S. pyogenes cells for DNA isolation are grown on half of the agar plate (Columbia with 5% sheep blood, BD, 221263; BioMerieux, 43 041).
(1) Scrape bacteria from the plate with the loop or swab.
(2) Re-suspend bacteria in 200 μl of TE buffer.
(3) Add 5 μl of RNase, 10 μl lysozyme and 2.5 μl of mutanolysin stocks.
(4) Incubate at 37°C for about 45 min.
(5) Follow with the standard chromosomal DNA isolation protocol.
(6) Dilute DNA to concentration ~10 ng/μl.
(7) Keep diluted template DNA at 4°C to avoid multiple freezing-thawing cycles.
Starters
Primer sequences were designed based on sequences of available S. pyogenes genomes. Amplicons sizes were designed so generated PCR products can be easily identified based on their size. We tested primer pairs individually so they are specific and yield only single PCR product. Primers for each PCR mix were tested in the multiplex PCR to test whether they behave the same way as individual primers.
(1) Mix equal volumes of individual 100 mM starters stock solutions into appropriate mixes, namely: “Mix 1,” “Mix 2,” “Mix 3” and “Mix 4.” Mixes 1 and 2 detect superantigens, Mix 3 detects DNases and Mix 4 detects proteases and sic gene. Primer sequences, composition of primer mixes and amplicon sizes within mixes are shown in
(2) Aliquot primers pre-mixes into 50–100 μl portions. (, Problem 1).
Table 2. Problem handling
Reaction setup
(1) All mixes, templates and reactions during setup should be kept on ice.
(2) Prepare diluted template DNA (concentration ~10 ng/μl). We preferably use 96 well format or 8/12 well strips to prepare templates. Using plate format noticeably speeds up the reaction setup. In majority of cases we run 96 well plate for each of four mixes and the same DNA template plate is used to setup all four plates.
(3) Mix together all reagents for PCR mastermixes according to . (, Problem 2).
Table 3. Reaction setup
(4) Dispense 2.2 μl (mastermix 1), 2.1 μl (mastermix 2 and 4) or 2.0 μl (mastermix 3) of the prepared mastermix to each well of the reaction plate or PCR tubes. Dispense mastermixes on the side of the tube/well. (, Problem 3).
(5) Using multichannel 0.5–10 μl pipette, add 2.8 μl of DNA template to wells/plates with mastermix 1, 2.9 μl to wells/plates with mastermixes 2 and 4, 3.0 μl to wells/plates with mastermix 3.
(6) Close tubes or seal the plate.
(7) Mix well and spin for 15 sec (tubes) or 1 min at 2500 rpm (plates).
(8) Place plate/tubes in a thermocycler and run program 1 or 2.
Equipment setup
The equipment does not need any special setup. We usually store pre-programmed PCR programs listed in .
Table 4. PCR program setup
Procedure
(1) PCR amplification
Amplification reactions are performed for 40 cycles using two different programs. Reactions with primer mixes 1–3 are amplified using program 1 and reactions with mix 4 are amplified using program 2.
(2) Gel electrophoresis
We analyze products of multiplex PCR reactions by electrophoresis in 1.5% (wt/vol) SeaKemLE agarose gel in TBE buffer, with addition ethidium bromide. Separation is performed with current 160 mV for about 100 min. Gels are photographed under UV light.
(3) Gel analysis
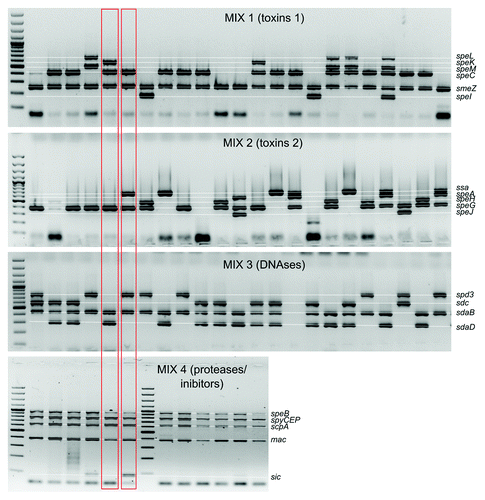
As a result of each multiplex reaction a set of multiple bands of various sizes can be observed (). Each PCR product corresponds with detected virulence factor. We usually note the absence/presence of a band as a 0 or 1 in a string of digits. As a result of four multiplex reactions we generate a string of 20 digits. Digitizing the results makes them easy to compare between strains. For example, during analysis of products generated with Mix1 code 000011 denotes that we did not detect genes speL, speK, speM and speC but detected products smeZ and speI. Example of the whole analysis can be seen on where marked strains would have codes 01011000010001111110 and 00011010010101011111.
Timing
The whole procedure can be performed during a single day, including DNA isolation.
(1) DNA isolation: ~90 min
(2) Reaction setup: ~15 min for 96 well plate, but only using multichannel pipettes and pre-dispensed template masterplate
(3) PCR amplification: Program 1, ~2 h 15 min; Program 2, ~4 h
(4) Gel electrophoresis: ~100 min
Anticipated Results
As a result of each multiplex reaction a set of multiple bands of various sizes can be observed. Presence of certain band denotes the presence of the particular gene. The generated strings of 20 digits can be used as a marker of each strain and be further used for strain clustering and the construction of phylogenetic trees. Categorization of strains based on the virulence factors correlates with emm typing and superantigens profile is a good predictor of emm type.Citation6
Primers from Mix 4 and sdaB primers from Mix 3 detect chromosomal genes that are present in virtually all GAS strain and they can serve as positive controls for PCR amplification. In our laboratory, we typed over 1,000 GAS strains and only in two cases we were unable to detect speB for unknown reasons. Products that correspond with sic gene are detected only in M1 serotype strains. Usually with M77 serotype strains we do not detect any genes from Mix2 and in some cases M81 strains do not yield any products after amplification with primers from Mix1. It is caused by lack of particular virulence factors in M77 and M81 strains, not by fault PCR amplification.
The method can be used when rapid strain comparison is required; it is fast and simple when compared with standard PFGE. The method is also inexpensive because it uses small reaction volumes (5 μl) and cost of reagents is markedly low.
The use of virulence factor detection can be used as an extension of any S. pyogenes typing and detection scheme. However if the researchers want to mix our primers with primers they use routinely, we recommend testing each individual primer pair they want to add to their typing scheme, whether it performs well in a mix. For laboratories that use method that detect 11 superantigens described by Lingtes,Citation5 we recommend use of Mix3 and Mix4 separately, as described in our manuscript to extend sensitivity and specificity of their detection.
Acknowledgments
The work was financed by the grant from National Center for Science (NCN) number NN401 536140.
References
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis 2005; 5:685 - 94; http://dx.doi.org/10.1016/S1473-3099(05)70267-X; PMID: 16253886
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 2000; 13:470 - 511; http://dx.doi.org/10.1128/CMR.13.3.470-511.2000; PMID: 10885988
- Proft T, Sriskandan S, Yang L, Fraser JD. Superantigens and streptococcal toxic shock syndrome. Emerg Infect Dis 2003; 9:1211 - 8; http://dx.doi.org/10.3201/eid0910.030042; PMID: 14609454
- Lintges M, van der Linden M, Hilgers RD, Arlt S, Al-Lahham A, Reinert RR, et al. Superantigen genes are more important than the emm type for the invasiveness of group A Streptococcus infection. J Infect Dis 2010; 202:20 - 8; http://dx.doi.org/10.1086/653082; PMID: 20497047
- Lintges M, Arlt S, Uciechowski P, Plümäkers B, Reinert RR, Al-Lahham A, et al. A new closed-tube multiplex real-time PCR to detect eleven superantigens of Streptococcus pyogenes identifies a strain without superantigen activity. Int J Med Microbiol 2007; 297:471 - 8; http://dx.doi.org/10.1016/j.ijmm.2007.03.015; PMID: 17481952
- Borek AL, Wilemska J, Izdebski R, Hryniewicz W, Sitkiewicz I. A new rapid and cost-effective method for detection of phages, ICEs and virulence factors encoded by Streptococcus pyogenes. Pol J Microbiol 2011; 60:187 - 201; PMID: 22184925