Abstract
For more than ten years the nematode Caenorhabditis elegans has proven to be a valuable model for studies of the host response to various bacterial and fungal pathogens. When exposed to a pathogenic organism, a clear response is elicited in the nematode, which is characterized by specific alterations on the transcriptional and translational levels. Early on, researchers took advantage of the possibility to conduct large-scale investigations of the C. elegans immune response. Multiple studies demonstrated that C. elegans does indeed mount a protective response against invading pathogens, thus rendering this small nematode a very useful and simple host model for the study of innate immunity and host-pathogen interactions. Here, we provide an overview of key aspects of innate immunity in C. elegans revealed by recent whole-genome transcriptomics and proteomics studies of the global response of C. elegans to various bacterial and fungal pathogens.
C. elegans as a Model Host
The nematode Caenorhabditis elegans was initially described as a model for bacterial infection in 1999Citation1,Citation2 and since then, it has proven to be an effective model host for several non-human and human pathogenic bacteria and fungi. In nature, C. elegans lives in rotting organic material and feeds on a variety of microorganisms and therefore, it continuously meets a diversity of bacterial and fungal pathogensCitation3 from which the worm needs to defend itself, making the nematode a valuable model for studying the host response in a whole organism context.
C. elegans lacks an adaptive immune system and it appears not to have specialized immune cells; however, infections trigger a clear response in the worm. C. elegans has become a valuable model host for the study of fundamental mechanisms in innate immunity and host-pathogen interactions, mainly due to its simplicity and ease of handling, and the wide range of experimental methods available for detailed studies in this organism. A hallmark of innate immunity is clearly present in the worm; a prominent defense response, including a battery of candidate immune effectors, is induced as C. elegans encounters various pathogens. Several immune signaling pathways have also been identified in the worm, and even though some host defense mechanisms in C. elegans have no homologs in the mammalian system, many pathways and effector molecules are conserved from worm to man.
In the laboratory, the nematode is routinely cultured on lawns of the non-pathogenic Escherichia coli strain OP50. Pathogenesis is easily evaluated in the worm by replacing the normal E. coli food source with the pathogen of interest and monitoring lifespan, or some other hallmark of the host response, like the expression of an immune effector gene. The normal median lifespan of C. elegans is 16–19 days on the standard E. coli food source, depending on the laboratory conditions. Different pathogens vary greatly in the time it takes for the worm to succumb to the bacterial infection; some pathogens kill within hours whereas the majority of pathogens used in laboratory experiments kill within 2 to 8 d.Citation4
The mode of bacterial infection varies from Yersenia pestis, which forms a biofilm that will attach to and cover the nematode mouth as it moves through the bacterial lawn, to Microbacterium nematophilum, which adheres to the hindgut cuticle; however, the majority of bacteria form an intestinal infection in C. elegans.Citation5 Fungi also vary in their mode of infection, as exemplified by Drechmeria coniospora that adheres to the cuticle and infects the nematode via its epidermis, and Harposporium sp and Candida albicans that infect C. elegans through its intestine.Citation6-Citation8 In recent studies the Gram-negative bacteria Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa were observed to escape from the intestinal lumen and invade the epithelial cells of the nematode.Citation9,Citation10 Furthermore, the microsporidium Nematocida parisii was shown to establish intracellular infections in the intestinal cells of C. elegans.Citation11 This pathogen, along with D. coniospora, is a natural pathogen of C. elegans, whereas the majority of pathogens used for C. elegans infection studies have not been observed infecting the worm outside of laboratory settings.
-Omics Studies of Innate Immunity
The C. elegans genome was the first completed animal genome and its completion opened up the way for large-scale investigations in this versatile biological model organism. Early on in the field of host-pathogen interactions, researchers took advantage of the possibility to gain genome-wide information of the innate immunity response in the worm, and several approaches have been used in the search for C. elegans immune defense genes or proteins. Microarrays were employed on worms infected by Serratia marcescens,Citation12 D. coniospora,Citation13 M. nematophilum,Citation14 P. aeruginosa,Citation15,Citation16 Staphylococcus aureus,Citation10 Y. pestis,Citation17 C. albicans,Citation8 Bacillus thuriengiensisCitation18 and a comparative study with S. marcescens, Enterococcus faecalis, Erwinia carotovora and Photorhabdus luminescens.Citation19 In addition to these transcriptional analyses of pathogen response, microarray was also performed to study the response to the pore-forming toxin Cry5B from B. thuringiensis.Citation20 Recently, tiling-arrays and RNA sequencing were used to study and compare the responses to S. marcescens, E. faecalis, P. luminescens, D. coniospora and Harposporium.Citation7 Studies focusing on the protein level during infection have also been performed, where the response to Aeromonas hydrophila and S. aureus, respectively, was investigated using differential gel electrophoresis,Citation21,Citation22 and infection with the pathogenic E. coli LF82 was analyzed using metabolic labeling and quantitative mass spectrometry.Citation23 A list of all -omics experiments targeting the C. elegans host response during infection can be seen in . These large-scale studies are in their nature very different since the pathogen, time points and methods vary between experiments. However, taken together, this large number of -omics studies hold a great deal of information on how C. elegans defends itself from a large variety of pathogens. When combining data from all -omics experiments, of all the genes/proteins identified in all the different large-scale experiments, 30% have only been found in one single experiment, suggesting that each pathogenic organism induces a specific immune response together with a more shared response.Citation7,Citation19 This review will focus on this shared portion of the C. elegans immune response.
Table 1. Genome-wide studies on the host response in C. elegans
C. elegans Core Set of Immune Effectors
In an attempt to define a core set of C. elegans immune regulators, we focused on genes/proteins, which were regulated (positively or negatively) in response to infection in eight or more experiments (out of the total 24 different large-scale infection experiments). This resulted in a set of 315 genes/proteins, of which 245 are mostly upregulated and 49 are mostly downregulated, whereas 21 genes/proteins are equally regulated (up or down) in at least eight experiments (Table S1). This core set is enriched in several protein families, domains and functional classes as can be seen in . The most common protein families identified in the core set are C-type lectins, DUF274 proteins, lysozymes, collagens and UDP-glucosyltransferases. The core data set is also enriched in CUB-domains and Metridin-like ShK toxin domains as well as a large number of genes/proteins with reported peptidase activity. These protein families will be described in detail below.
Figure 1. The core C. elegans immune response is enriched in several protein families and domains. Pie chart of the protein classes enriched in the core immune host response defined as genes/proteins being regulated in eight or more genome-wide experiments of the C. elegans response to different pathogens. The full list of genome-wide experiments can be seen in and a list of the core response genes/proteins is found Table S1. The enrichment analysis was performed using DAVID v. 6.7 (http://david.abcc.ncifcrf.gov/).Citation71
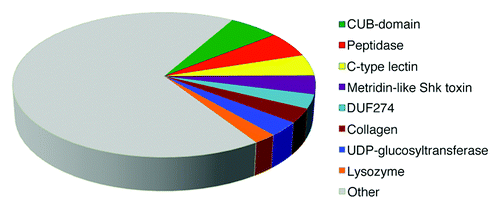
Several genes/proteins are found to be exclusively upregulated in the 24 -omics studies and curiously, very little is known about the immune regulators exclusively upregulated in half or more studies: The F53A9.1, F53A9.2 and C50F7.5 genes encode completely uncharacterized proteins, and oac-31 encodes a O-Acyltransferase homolog. Also among the most often upregulated genes are cdr-4, a cadmium responsive gene; and clc-1, a claudin homolog, which is an important component of tight junctions and believed to maintain the impermeability of the epithelial layer. clc-1 was shown to be required for resistance toward S. aureus in the nematode.Citation24
Interestingly, no genes/proteins are found to be exclusively downregulated; however, several genes are found to be much more often downregulated than upregulated as a response to a pathogen. Among these are vit-1 and vit-3, two vittelogenins, the major egg yolk proteins in C. elegans, as well as the two C. elegans metallothioneins mtl-1 and mtl-2. Metallothioneins are small, cysteine-rich metal-ion binding proteins, which serve roles in cadmium toxicity protection, zinc homeostasis, and even a role in immunosuppression has been found.Citation25,Citation26 Their role, if any, in the C. elegans immune response remains unknown. The downregulated core set also includes cnc-2, which encodes an antimicrobial peptide of the caenacin family known to promote survival of C. elegans after fungal infection,Citation27 and thn-2 encoding an antimicrobial protein of the thaumatin family conferring resistance to infection by pathogens such as P. aeruginosaCitation28 and M. nematophilum.Citation14 At first glance, downregulation of host defense effectors in response to infection may appear rather surprising. However, in some cases this may reflect a mechanism by which pathogens overcome the immune defenses of C. elegans, as demonstrated for P. aeruginosa.Citation28 The presence of several immune defense effectors among the core set of genes mostly downregulated upon infection suggests that suppression of host immunity is a general strategy employed by pathogens to resist C. elegans host defenses.
C-Type Lectins
In C. elegans, C-type lectins have been reported in the response to almost every single pathogen tested, and the total number of different C-type lectins induced is over 60.Citation7,Citation10,Citation14-Citation16,Citation19,Citation22,Citation23,Citation28,Citation29 Thirteen C-type lectins and an additional two lectins are found in the core set of C. elegans immune regulators. C-type lectins belong to a large superfamily (> 200 proteins) containing C-type lectin-like domains (CTLDs). A classical C-type lectin binds carbohydrate structures in a Ca2+-dependent manner, but many CTLDs bind carbohydrates independently of Ca2+ and some bind lipids or other ligands.Citation30 The domain architecture of the vertebrate CTLDs differ from the invertebrates; the only common architecture found is a single CTLD, which is also the most common architecture in C. elegans.Citation31 The differential pattern exhibited by C-type lectins to different pathogens was proposed as a potential mechanism for the innate immune specificity observed in C. elegans,Citation32 but clearly, a part of the C-type lectins are induced in response to several different pathogens. Interestingly, of the C-type lectins found in the core set, the two fungi C. albicans and D. coniospora seem to mostly downregulate the expression of these proteins, whereas the majority of the other pathogens seem to upregulate them. Only one C-type lectin (clec-61) is more often downregulated than upregulated. A subset of C. elegans C-type lectins have been tested directly with respect to their effect on survival of the host, and clec-17, clec-60 and clec-86 were reported to cause enhanced susceptibility to M. nematophilum when knocked down by RNAi.Citation14 Also, knockdown of clec-70 resulted in enhanced susceptibility to S. aureus,Citation10 and simultaneous overexpression of clec-60 and clec-61, or clec-70 and clec-71 resulted in enhanced resistance to S. aureus but, interestingly, decreased resistance to P. aeruginosa.Citation10 RNAi knockdown of clec-65 also caused enhanced susceptibility to the E. coli LF82.Citation23 In mammals, the C-type lectins mouse RegIIIγ and human HIP/PAP were shown to be induced by intestinal bacteria and to bind pepdidoglycan, and they possess direct antimicrobial activity against Gram-positive bacteria.Citation33 However, so far, no functional studies can confirm whether any of the C. elegans C-type lectins share this function with their mammalian counterparts.
CUB Domains
The core set is highly enriched in proteins containing CUB (or CUB-like) domains (C1r/C1s, Uegf and Bmp1). More than 50 CUB-like proteins are encoded from the C. elegans genome. The CUB domain is a structural motif of approximately 110 residues found almost exclusively in extracellular and plasma membrane-associated proteins with a diverse range of functions. It has been suggested that C. elegans CUB-domain proteins function in innate immunity, due to the organization of their genes in large clusters and the similarity of CUB domains to immunoglobulins.Citation34 Indeed, functional roles in pathogen resistance have been demonstrated for several of the 18 CUB-like genes belonging to the core set of upregulated genes. Inhibition of F08G5.6 enhanced the susceptibility of C. elegans toward both P. aeruginosaCitation15,Citation35 and Y. pestis.Citation17 Knockdown of F20G2.5 and dct-17 also led to an increased sensitivity to P. aeruginosa,Citation15,Citation35 whereas RNAi inhibition of C17H12.8 and C32H11.12 enhanced the susceptibility of C. elegans to Y. pestis infection.Citation17 Curiously, the fungi C. albicans and D. coniospora appear to downregulate several CUB-like genes mostly upregulated by other pathogens (see Table S1).
DUF274 Proteins
The C. elegans genome encodes 22 proteins with the domain of unknown function 274 (DUF274); a gene family unique to Caenorhabditis. A remarkable ten DUF274 proteins are part of the core immune response, yet very little functional information is available for this protein family, in which all members consist solely of the DUF274 domain. Only one study has shown a role for a DUF274 protein; RNAi knockdown of Y41D4B.16 significantly reduced the lifespan on the pathogenic E. coli LF82,Citation23 but the very strong presence of this protein family in the core set suggests an important function for the DUF274 proteins in the C. elegans immune response. Interestingly, the fungus D. coniospora seems to downregulate expression of the DUF274 proteins, whereas the rest of the pathogens almost exclusively upregulate these proteins.
UDP-Glucuronosyltransferases and Metridin-Like ShK Toxin Domains
The C. elegans core immune response is enriched in enzymes, which transfer a glycosyl group from an UTP-sugar to a small hydrophobic molecule, the UDP-glucuronosyltransferases (UGTs). This glucuronidation reaction is an important detoxification process in vertebrates, however, UDP-glucuronosyltransferases (together with cytochrome P450s and gluthathione S-transferases, of which cyp-25A2, cyp-34A9, cyp-35C1, gst-20 and gst-38 are also in the core response) have also been associated with olfactory processing.Citation36 Little functional information is available for the UGTs in C. elegans, but their presence in the core response is intriguing, since C. elegans has been shown to exhibit olfactory learning against pathogenic bacteria,Citation37-Citation39 but if this involves the UGTs is yet to be determined. Moreover, functional annotation of the UGTs implies that this class of enzymes could be involved in transfer of a glycosyl group to lipids, suggesting that lipid modification is involved in the core host response against pathogens. Previously, it was suggested that C. elegans resists the toxic Cry5B produced by B. thuringiensis by making specific glycolipid-binding galectins, which compete out and hence prevent Cry5B binding in the intestine of C. elegans.Citation40
Metridin-like ShK toxin domains have a structural resemblance to ShK toxin produced by sea anemones, which is known to block potassium channels, however, the metridin-like ShK toxin domain lacks a lysine residue necessary for this function.Citation41 The functional relevance of this protein domain in the C. elegans immune response is completely unknown.
Lysozymes
Lysozymes are antimicrobial proteins present in microbes, insects, plants and animals. They cause lysis of bacterial cells by breaking down peptidoglycan, the major component of the bacterial cell wall. C. elegans has ten genes encoding lysozymes (lys-1 to lys-10), which belong to the protist-type lysozymes, and an additional five invertebrate-type lysozymes (ilys-1 to ilys-5).Citation42 The lysozymes have been studied with regards to many different infections in C. elegans. It is therefore to be expected that several lysozymes are part of the C. elegans core immune response. Five lysozymes are more often seen to be upregulated in response to pathogenic infection (lys-1, -2, -4, -6 and ilys-2); one (lys-5) is regulated equally up and down, whereas ilys-3 is more often downregulated in response to infection. Overexpression of lys-1 made C. elegans more resistant to a specific strain of S. marcescens,Citation12 and knockdown of lys-1 caused enhanced susceptibility to S. aureus.Citation24 Simultaneous overexpression of the genomic neighbors lys-4 and lys-5 caused resistance to S. aureusCitation10 while the loss of lys-7 resulted in less resistance to M. nematophilum and the pathogenic E. coli LF82.Citation14,Citation23 Knockdown of lys-7 also caused enhanced susceptibility toward P. aeruginosa and, interestingly, this pathogen suppresses the expression of lys-7 as part of its pathogenicity strategy.Citation28 In contrast, more resistance toward S. Typhimurium was observed in a lys-7 mutant, which was also shown to be more susceptible to infection with the fungus Cryptococcus neoformans.Citation43 Furthermore, knockdown of lys-2, lys-5 and lys-7 led to decreased resistance against B. thuringiensis, whereas overexpression of lys-5 and lys-7, but not lys-2, resulted in an increase in resistance.Citation18 The direct mode of action of any lysozyme has not been confirmed in C. elegans, but their large representation in the core set of immune effectors suggests an important function in the defense system of the worm.
Collagens
Collagens, which are the major part of the C. elegans exoskeleton called the cuticle, are also among the highly regulated genes/proteins in the host response. The majority (nine out of ten) are more often upregulated in response to an invading pathogen, and the trend seems to be that the two fungi Harposporium and D. coniospora strongly downregulate the expression of numerous collagens, whereas the bacterial pathogens are found to both up- and downregulate the collagens. The collagenous cuticle is an important line of defense for C. elegans, and with D. coniospora infecting via the cuticle, it comes as no surprise that this pathogen would potentially target the collagens. However, Harposporium, and the many bacterial pathogens observed to regulate collagens, all infect via the intestine, and the role of the collagens here is less obvious. The collagen genes in C. elegans have highly similar nucleotide sequences and thus cross-hybridization is likely to occur on the microarrays, so when examining this group of genes, this potential error needs to be taken into account.
Signaling in the Immune Response
Several signaling cascades involved in the response to pathogenic attack have been identified in C. elegans and importantly, the major pathways have all been shown to play important roles in mammalian innate immunity as well.Citation44 Key components in the nematode host response include the p38 mitogen-activated protein kinase (MAPK), the transforming growth factor-β (TGFβ) and the insulin-like DAF-2-DAF-16 pathways, but also the bZIP transcription factor ZIP-2 is involved in responding to pathogens. Interestingly, the Toll-like receptor (TLR), which has a very central role in mammalian innate immunity, does not seem to be an important component of the immune response of the worm, since partial loss of function in the single TLR homolog in C. elegans, tol-1, does not affect the defense against the majority of pathogens tested, albeit an enhanced susceptibility to infection by S. enterica has been reported.Citation44-Citation47 Insights learned from C. elegans could therefore not only give novel information on the fundamental immune pathways conserved in humans and nematodes; the nematode might also present an opportunity to identify TLR-independent mechanisms of innate immunity.
The core set of immune effectors identified from the large-scale infection experiments in C. elegans does indeed suggest that the known signaling pathways are very important in the shared host response. Several immune effectors in the core immune response are known to be regulated by the MAPK pathway in C. elegans, including lys-1, lys-2, three DUF274 proteins and several CUB-like proteins.Citation16 The conserved p38 MAPK pathway includes the two upstream kinases NSY-1, SEK-1 and the MAPK PMK-1, which cause enhanced susceptibility to P. aeruginosa when mutated.Citation48 One of the transcription factors downstream of PMK-1 is ATF-7, and lack of this conserved transcription factor causes hyper-susceptibility to some pathogens.Citation49 A similar p38 MAPK cascade also plays an important role in innate immunity in the mammalian system, where it is present downstream of the TLR, but as mentioned above, the single TLR in the nematode is not central in the innate immune response for C. elegans.Citation44-Citation47 In addition to tol-1, C. elegans tir-1 contains a Toll/IL-1R homology (TIR) domain; the cytoplasmic signaling domain of the mammalian TLR. tir-1 was shown to function upstream of NSY-1 in the p38 MAPK cascade,Citation47 and mutations or knockdown in tir-1 resulted in susceptibility to a variety of pathogens.Citation47,Citation50 The upstream components of the MAPK cascade differ depending on the tissue. In the intestine, the protein kinase Cδ (PKCδ) TPA-1 activates the protein kinase D DKF-2, which in turn activates the PMK-I cassette.Citation51 Lack of DKF-2 caused less resistance to P. aeruginosa and E. faecalis, and inactivation of TPA-1 also caused less resistance to P. aeruginosa. In the hypodermis, the PKCδ TPA-1 is also involved in this signaling, and a study identified two PLCs, EGL-8 and PLC-3, and the two proteins Gα GPA-12 and Gβ RACK-1, as additional upstream components.Citation52
The core immune response also comprises several known targets of the TGFβ-signaling pathway, including four C-type lectins and irg-2.Citation53 In C. elegans, DBL-1 (a TGF-β homolog) binds to the DAF-4/SMA-6 receptor and acts through the SMAD complex. dbl-1 mutants are hypersensitive to infection with S. marcescens,Citation12 and the dbl-1 gene was also shown to be necessary for the expression of cnc-2 after D. coniospora infection.Citation27 In the epidermis, the smad proteins SMA-2 and SMA-4 were not necessary for the induction of cnc-2.
The FOXO transcription factor DAF-16 regulates several genes involved in stress-response, longevity and innate immunity in C. elegans,Citation54-Citation56 including eight genes named on the basis of their regulation by DAF-16 (dct-7, -16, -17 and -18 and dod-3, -17, -19 and -22). The DAF-2 insulin/IGF-1-like receptor inhibits DAF-16 by phosphorylation via a kinase cascade, including AGE-1, PDK-1, AKT-1, AKT-2 and SGK-1. In DAF-2 mutants, DAF-16 is constitutively active, and pathogen-resistance of daf-2 mutants has been observed with numerous pathogens; this phenotype is DAF-16 dependent.Citation16,Citation23,Citation57,Citation58 The DAF-2-DAF-16 pathway also regulates lifespan, and the daf-2 mutants are long lived in addition to being resistant to pathogens. These observations could be two alternative descriptions of the same phenomena; however, this is not the case, since regulation of lifespan and pathogen resistance can function independently,Citation59 and different components of the insulin-like signaling pathway distinctly regulate lifespan and pathogen resistance. Along with DAF-2, AGE-1, AKT-1 and AKT-2 regulate both lifespan and resistance, whereas SGK-1 and PDK-1 regulate lifespan but have no effect on pathogen resistance.
Defense against P. aeruginosa is also mediated by the bZIP transcription factor ZIP-2. P. aeruginosa infection involves the action of exotoxin A, which causes translation inhibition; this blockade of host protein translation will in turn activate the ZIP-2 pathway and several immune genes are consequently transcribed.Citation60-Citation62 Curiously, ZIP-2 affects the expression of genes belonging to the core host response, including irg-2 and Y58A7A.5, suggesting that ZIP-2 could be involved in defense responses to more pathogens in addition to P. aeruginosa. This is very interesting since ZIP-2 signaling has been speculated to be used by the C. elegans host to discriminate between non-pathogenic bacteria and pathogenic bacteria causing disruption in cellular homeostasis.Citation61-Citation64 ZIP-2 has no direct homolog in mammals; however the bZIP transcription factor class has been shown to be involved in mammalian innate immunity.Citation60
Variance between Large-Scale Investigations
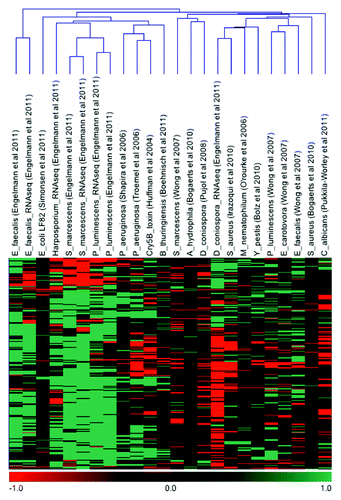
A hierarchical clustering analysis of the core immune response reveals that responses of different pathogens show varying degrees of similarity among each other (). Especially the response toward D. coniospora seems to be mainly oppositely regulated; however, this fungus also infects C. elegans differently than the majority of the other pathogens by adhering to the cuticle of the worm and then infecting via the epidermis. To a lesser degree, the response reported toward C. albicans is also largely opposite to the bacterial pathogens tested, even though this fungus infects via the intestine, so it would seem that the worm responds differently toward fungi than to bacterial pathogens. Interestingly, the two fungi seem to mainly downregulate genes, whereas the majority of bacterial pathogens induce upregulation of a much larger number of genes than they downregulate. To date, the large-scale investigations of C. elegans immunity show large variance, even for studies of responses to the same pathogen; e.g., two different investigations of S. marcescens infection proved less than 2% overlap, and in the case of different studies on P. aeruginosa infection, the overlap was less than 20%.Citation19 Data from microarray and proteomics investigations of S. aureus infection differs even more; only five proteins of the 108 proteins reported as differentially regulated in the proteomics study were also seen regulated on the transcriptional level.Citation10,Citation21 Despite these large differences, an increasingly more thorough picture of C. elegans immunity is emerging based on the accumulation of all these observations. By focusing on genes/proteins only expressed in eight or more -omics experiments, a common set of immune modulators was defined, including well known immune effectors, such as the C-type lectins, lysozymes and CUB-domain proteins, which have all been described as induced by the majority of pathogens investigated. Additionally, our core set of immune effectors also shows how collagens and UDP-glucuronosyltransferase are very common in the C. elegans host response.
Figure 2. Hierarchical clustering analysis of the core immune response in C. elegans. The clustering analysis was performed using the software MeV v4.8. Green indicates induction of the corresponding gene/protein and red indicates repression under the designated condition.
The C. elegans immunity response shares features of other stress responses such as heat shock, osmotic, ER (endoplasmatic reticulum) and heavy metal stress,Citation20,Citation65-Citation67 but since an infection causes large cellular and eventually organismal damage, it seems only natural that many stress responses overlap. Importantly, a large pool of large scale data is now available for the innate immune response of the worm. This opens up for the comparison with this data to other biological responses, as has been exemplified in several studies, including analyses of miRISC-associated mRNAs (mRNAs) in the intestine, and animals containing mutations in the DICER gene, respectively, demonstrating a significant overlap with pathogen-responsive mRNAs.Citation68,Citation69
Future Directions
The techniques for performing -omics experiment are constantly improving and as such the precision and quality of data will as well. The majority of experiments have been performed using microarray, but with the recent addition of RNA-sequencing employed to deduce the host response, an unprecedented quantitative measure of transcript levels can be obtained. So far, the transcriptional responses to invading pathogens were investigated in far greater detail than the translational response. However, with the recent elegant approach enabling stable-isotope labeling with amino acids in cell culture (SILAC) in C. elegans by feeding worms a heavy lysine- and heavy arginine-labeled E. coli strain, resulting in very precise and deep investigation of the proteome,Citation70 quantitative proteomics should in the future be used to gain further information on the infection response on the protein level. The complex host-pathogen relationship could also benefit from simultaneous large-scale investigation of the response of the host as well as the pathogen. To our knowledge, such experiments have not been performed in C. elegans. Future -omics investigation in C. elegans will help give more knowledge on the host response to various pathogens, but one major concept is already clear; C. elegans is clearly capable of mounting both pathogen-specific and pathogen-shared responses to an infection.
Additional material
Download Zip (68 KB)Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/virulence/article/21906
References
- Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 1999; 96:47 - 56; http://dx.doi.org/10.1016/S0092-8674(00)80958-7; PMID: 9989496
- Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 1999; 96:2408 - 13; http://dx.doi.org/10.1073/pnas.96.5.2408; PMID: 10051655
- Félix MA, Braendle C. The natural history of Caenorhabditis elegans. Curr Biol 2010; 20:R965 - 9; http://dx.doi.org/10.1016/j.cub.2010.09.050; PMID: 21093785
- Sifri CD, Begun J, Ausubel FM. The worm has turned--microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol 2005; 13:119 - 27; http://dx.doi.org/10.1016/j.tim.2005.01.003; PMID: 15737730
- Nicholas HR, Hodgkin J. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol Immunol 2004; 41:479 - 93; http://dx.doi.org/10.1016/j.molimm.2004.03.037; PMID: 15183927
- Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 2009; 8:1750 - 8; http://dx.doi.org/10.1128/EC.00163-09; PMID: 19666778
- Engelmann I, Griffon A, Tichit L, Montañana-Sanchis F, Wang G, Reinke V, et al. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS One 2011; 6:e19055; http://dx.doi.org/10.1371/journal.pone.0019055; PMID: 21602919
- Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog 2011; 7:e1002074; http://dx.doi.org/10.1371/journal.ppat.1002074; PMID: 21731485
- Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A 2009; 106:14564 - 9; PMID: 19667176
- Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog 2010; 6:e1000982; http://dx.doi.org/10.1371/journal.ppat.1000982; PMID: 20617181
- Troemel ER, Félix MA, Whiteman NK, Barrière A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol 2008; 6:2736 - 52; http://dx.doi.org/10.1371/journal.pbio.0060309; PMID: 19071962
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol 2002; 12:1209 - 14; http://dx.doi.org/10.1016/S0960-9822(02)00928-4; PMID: 12176330
- Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, et al. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol 2008; 18:481 - 9; http://dx.doi.org/10.1016/j.cub.2008.02.079; PMID: 18394898
- O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res 2006; 16:1005 - 16; http://dx.doi.org/10.1101/gr.50823006; PMID: 16809667
- Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A 2006; 103:14086 - 91; http://dx.doi.org/10.1073/pnas.0603424103; PMID: 16968778
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2006; 2:e183; http://dx.doi.org/10.1371/journal.pgen.0020183; PMID: 17096597
- Bolz DD, Tenor JL, Aballay A. A conserved PMK-1/p38 MAPK is required in caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J Biol Chem 2010; 285:10832 - 40; http://dx.doi.org/10.1074/jbc.M109.091629; PMID: 20133945
- Boehnisch C, Wong D, Habig M, Isermann K, Michiels NK, Roeder T, et al. Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis. PLoS One 2011; 6:e24619; http://dx.doi.org/10.1371/journal.pone.0024619; PMID: 21931778
- Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol 2007; 8:R194; http://dx.doi.org/10.1186/gb-2007-8-9-r194; PMID: 17875205
- Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, Aroian RV. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A 2004; 101:10995 - 1000; http://dx.doi.org/10.1073/pnas.0404073101; PMID: 15256590
- Bogaerts A, Beets I, Temmerman L, Schoofs L, Verleyen P. Proteome changes of Caenorhabditis elegans upon a Staphylococcus aureus infection. Biol Direct 2010; 5:11; http://dx.doi.org/10.1186/1745-6150-5-11; PMID: 20163716
- Bogaerts A, Temmerman L, Boerjan B, Husson SJ, Schoofs L, Verleyen P. A differential proteomics study of Caenorhabditis elegans infected with Aeromonas hydrophila. Dev Comp Immunol 2010; 34:690 - 8; http://dx.doi.org/10.1016/j.dci.2010.02.003; PMID: 20149819
- Simonsen KT, Møller-Jensen J, Kristensen AR, Andersen JS, Riddle DL, Kallipolitis BH. Quantitative proteomics identifies ferritin in the innate immune response of C. elegans. Virulence 2011; 2:120 - 30; http://dx.doi.org/10.4161/viru.2.2.15270; PMID: 21389771
- Jensen VL, Simonsen KT, Lee YH, Park D, Riddle DL. RNAi screen of DAF-16/FOXO target genes in C. elegans links pathogenesis and dauer formation. PLoS One 2010; 5:e15902; http://dx.doi.org/10.1371/journal.pone.0015902; PMID: 21209831
- Zeitoun-Ghandour S, Charnock JM, Hodson ME, Leszczyszyn OI, Blindauer CA, Stürzenbaum SR. The two Caenorhabditis elegans metallothioneins (CeMT-1 and CeMT-2) discriminate between essential zinc and toxic cadmium. FEBS J 2010; 277:2531 - 42; http://dx.doi.org/10.1111/j.1742-4658.2010.07667.x; PMID: 20553489
- Canpolat E, Lynes MA. In vivo manipulation of endogenous metallothionein with a monoclonal antibody enhances a T-dependent humoral immune response. Toxicol Sci 2001; 62:61 - 70; http://dx.doi.org/10.1093/toxsci/62.1.61; PMID: 11399794
- Zugasti O, Ewbank JJ. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-beta signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol 2009; 10:249 - 56; http://dx.doi.org/10.1038/ni.1700; PMID: 19198592
- Evans EA, Kawli T, Tan MW. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog 2008; 4:e1000175; http://dx.doi.org/10.1371/journal.ppat.1000175; PMID: 18927620
- Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol 2007; 27:5544 - 53; http://dx.doi.org/10.1128/MCB.02070-06; PMID: 17526726
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J 2005; 272:6179 - 217; http://dx.doi.org/10.1111/j.1742-4658.2005.05031.x; PMID: 16336259
- Drickamer K, Dodd RB. C-Type lectin-like domains in Caenorhabditis elegans: predictions from the complete genome sequence. Glycobiology 1999; 9:1357 - 69; http://dx.doi.org/10.1093/glycob/9.12.1357; PMID: 10561461
- Schulenburg H, Hoeppner MP, Weiner J 3rd, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 2008; 213:237 - 50; http://dx.doi.org/10.1016/j.imbio.2007.12.004; PMID: 18406370
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006; 313:1126 - 30; http://dx.doi.org/10.1126/science.1127119; PMID: 16931762
- Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol 1993; 231:539 - 45; http://dx.doi.org/10.1006/jmbi.1993.1305; PMID: 8510165
- Nandakumar M, Tan MW. Gamma-linolenic and stearidonic acids are required for basal immunity in Caenorhabditis elegans through their effects on p38 MAP kinase activity. PLoS Genet 2008; 4:e1000273; http://dx.doi.org/10.1371/journal.pgen.1000273; PMID: 19023415
- Wang Q, Hasan G, Pikielny CW. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J Biol Chem 1999; 274:10309 - 15; http://dx.doi.org/10.1074/jbc.274.15.10309; PMID: 10187818
- Beale E, Li G, Tan MW, Rumbaugh KP. Caenorhabditis elegans senses bacterial autoinducers. Appl Environ Microbiol 2006; 72:5135 - 7; http://dx.doi.org/10.1128/AEM.00611-06; PMID: 16820523
- Rumbaugh KP. Fatal attraction: bacterial bait lures worms to their death. Proc Natl Acad Sci U S A 2010; 107:16411 - 2; http://dx.doi.org/10.1073/pnas.1011935107; PMID: 20823245
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 2005; 438:179 - 84; http://dx.doi.org/10.1038/nature04216; PMID: 16281027
- Ideo H, Fukushima K, Gengyo-Ando K, Mitani S, Dejima K, Nomura K, et al. A Caenorhabditis elegans glycolipid-binding galectin functions in host defense against bacterial infection. J Biol Chem 2009; 284:26493 - 501; http://dx.doi.org/10.1074/jbc.M109.038257; PMID: 19635802
- Shivers RP, Youngman MJ, Kim DH. Transcriptional responses to pathogens in Caenorhabditis elegans. Curr Opin Microbiol 2008; 11:251 - 6; http://dx.doi.org/10.1016/j.mib.2008.05.014; PMID: 18567532
- Schulenburg H, Boehnisch C. Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae). BMC Evol Biol 2008; 8:114; http://dx.doi.org/10.1186/1471-2148-8-114; PMID: 18423043
- Marsh EK, van den Berg MC, May RC. A two-gene balance regulates Salmonella typhimurium tolerance in the nematode Caenorhabditis elegans. PLoS One 2011; 6:e16839; http://dx.doi.org/10.1371/journal.pone.0016839; PMID: 21399680
- Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol 2010; 10:47 - 58; http://dx.doi.org/10.1038/nri2689; PMID: 20029447
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol 2001; 11:809 - 21; http://dx.doi.org/10.1016/S0960-9822(01)00241-X; PMID: 11516642
- Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep 2008; 9:103 - 9; http://dx.doi.org/10.1038/sj.embor.7401104; PMID: 17975555
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol 2004; 5:488 - 94; http://dx.doi.org/10.1038/ni1060; PMID: 15048112
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 2002; 297:623 - 6; http://dx.doi.org/10.1126/science.1073759; PMID: 12142542
- Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, et al. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet 2010; 6:e1000892; http://dx.doi.org/10.1371/journal.pgen.1000892; PMID: 20369020
- Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A 2004; 101:6593 - 8; http://dx.doi.org/10.1073/pnas.0308625101; PMID: 15123841
- Ren M, Feng H, Fu Y, Land M, Rubin CS. Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity 2009; 30:521 - 32; http://dx.doi.org/10.1016/j.immuni.2009.03.007; PMID: 19371715
- Ziegler K, Kurz CL, Cypowyj S, Couillault C, Pophillat M, Pujol N, et al. Antifungal innate immunity in C. elegans: PKCdelta links G protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe 2009; 5:341 - 52; http://dx.doi.org/10.1016/j.chom.2009.03.006; PMID: 19380113
- Roberts AF, Gumienny TL, Gleason RJ, Wang H, Padgett RW. Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans. BMC Dev Biol 2010; 10:61; http://dx.doi.org/10.1186/1471-213X-10-61; PMID: 20529267
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, et al. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 2007; 317:660 - 3; http://dx.doi.org/10.1126/science.1139952; PMID: 17673661
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003; 424:277 - 83; http://dx.doi.org/10.1038/nature01789; PMID: 12845331
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 2006; 38:251 - 7; http://dx.doi.org/10.1038/ng1723; PMID: 16380712
- Chen CS, Bellier A, Kao CY, Yang YL, Chen HD, Los FC, et al. WWP-1 is a novel modulator of the DAF-2 insulin-like signaling network involved in pore-forming toxin cellular defenses in Caenorhabditis elegans. PLoS One 2010; 5:e9494; http://dx.doi.org/10.1371/journal.pone.0009494; PMID: 20209166
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 2003; 300:1921; http://dx.doi.org/10.1126/science.1080147; PMID: 12817143
- Evans EA, Chen WC, Tan MW. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell 2008; 7:879 - 93; http://dx.doi.org/10.1111/j.1474-9726.2008.00435.x; PMID: 18782349
- Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2010; 107:2153 - 8; http://dx.doi.org/10.1073/pnas.0914643107; PMID: 20133860
- Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 2012; 11:375 - 86; http://dx.doi.org/10.1016/j.chom.2012.02.008; PMID: 22520465
- McEwan DL, Kirienko NV, Ausubel FM. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 2012; 11:364 - 74; http://dx.doi.org/10.1016/j.chom.2012.02.007; PMID: 22520464
- Kleino A, Silverman N. UnZIPping mechanisms of effector-triggered immunity in animals. Cell Host Microbe 2012; 11:320 - 2; http://dx.doi.org/10.1016/j.chom.2012.04.002; PMID: 22520459
- Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 2012; 149:452 - 66; http://dx.doi.org/10.1016/j.cell.2012.02.050; PMID: 22500807
- Mohri-Shiomi A, Garsin DA. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J Biol Chem 2008; 283:194 - 201; http://dx.doi.org/10.1074/jbc.M707956200; PMID: 17951251
- Rohlfing AK, Miteva Y, Hannenhalli S, Lamitina T. Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS One 2010; 5:e9010; http://dx.doi.org/10.1371/journal.pone.0009010; PMID: 20126308
- Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci U S A 2006; 103:13092 - 7; http://dx.doi.org/10.1073/pnas.0604050103; PMID: 16916933
- Kudlow BA, Zhang L, Han M. Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol Cell 2012; 46:530 - 41; http://dx.doi.org/10.1016/j.molcel.2012.03.011; PMID: 22503424
- Welker NC, Habig JW, Bass BL. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA 2007; 13:1090 - 102; http://dx.doi.org/10.1261/rna.542107; PMID: 17526642
- Larance M, Bailly AP, Pourkarimi E, Hay RT, Buchanan G, Coulthurst S, et al. Stable-isotope labeling with amino acids in nematodes. Nat Methods 2011; 8:849 - 51; http://dx.doi.org/10.1038/nmeth.1679; PMID: 21874007
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44 - 57; http://dx.doi.org/10.1038/nprot.2008.211; PMID: 19131956