Abstract
The genetic similarity between Burkholderia mallei (glanders) and Burkholderia pseudomallei (melioidosis) had led to the general assumption that pathogenesis of each bacterium would be similar. In 2000, the first human case of glanders in North America since 1945 was reported in a microbiology laboratory worker. Leveraging the availability of pre-exposure sera for this individual and employing the same well-characterized protein array platform that has been previously used to study a large cohort of melioidosis patients in southeast Asia, we describe the antibody response in a human with glanders. Analysis of 156 peptides present on the array revealed antibodies against 17 peptides with a > 2-fold increase in this infection. Unexpectedly, when the glanders data were compared with a previous data set from B. pseudomallei infections, there were only two highly increased antibodies shared between these two infections. These findings have implications in the diagnosis and treatment of B. mallei and B. pseudomallei infections.
Burkholderia mallei and the closely related Burkholderia pseudomallei are CDC Category B Bioterrorism Agents due to the history of confirmed use of B. mallei in biological warfare including the US Civil War,Citation1 World War I,Citation2 World War IICitation1,Citation3 and purportedly in Afghanistan in the 1980s.Citation4B. mallei is an obligate pathogen of horses that causes glanders, a chronic disease known since the time of Aristotle,Citation1 that can infect humans who work in close proximity to infected animals.Citation1,Citation5 In this work we employ a protein microarray, which was previously used in the study of a large cohort of patients in southeast Asia with B. pseudomallei infections,Citation6 to analyze the targets of antibodies produced against B. mallei in the first human case of glanders in the US since 1946.Citation7,Citation8 This work provides the first direct comparison of the human antibody reaction against B. mallei and against B. pseudomallei. Importantly, despite the high level of similarity between B. mallei and B. pseudomallei and the similarity in disease presentation, the antibody profiles are strikingly different. This suggests that different therapeutic approaches might be required for each infection and also provides potential antigens for the development of a practical differential diagnosis approach.
Glanders has been eradicated from most of Europe and all of North America through aggressive infection control programs.Citation1 As a result little is known about Burkholderia mallei pathogenesis in humans compared with Burkholderia pseudomallei, a related environmental bacterium and opportunistic pathogen, endemic in southeast Asia.Citation5
In 2000, a worker at United States Army Medical Research Institute of Infectious Diseases (USAMRIID) accidentally acquired a B. mallei infection.Citation8 This case has been the subject of previous reports due in part to the unique opportunity to study a human glanders infection for which pre-exposure and post-exposure serum exists.Citation9,Citation10 A recent analysis indicated that levels of B. mallei-specific IgA, IgG and IgM were highly elevated at 64 d post-infection,Citation10 but the immunogenic antigens were not identified. We employed a previously described B. pseudomallei protein arrayCitation6,Citation11 to perform an in-depth analysis of this serum. This array was previously used to identify antibodies produced against B. pseudomallei in a cohort of melioidosis patients in southeast Asia.Citation6,Citation11 The protein microarray incorporates 214 B. pseudomallei K96243 computationally-predicted antigenic peptides, and construction of this array was previously described.Citation6 The B. mallei genome is a reduced version of the B. pseudomallei genome that has 99.1% identity for shared genes and does not contain additional genes.Citation5 Accordingly, the B. pseudomallei protein microarray can be used to detect reactivity to B. mallei proteins as 156 of the peptides are present in some form in both species (Table S1).Citation12,Citation13 Microarrays were hybridized using pre-exposure serum and serum from 2 mo after symptoms manifested in the researcher who had contracted glanders.Citation8 Hybridization, image scanning, and data acquisition were performed as previously described.Citation6 Data were analyzed by generating log2 ratios of (post-exposure intensity/pre-exposure intensity).
When compared with the pre-exposure serum, the log2 ratio of post-exposure to pre-exposure intensities were > 2 for 7 out of 156 peptides present on the array and between 1 and 2 for 12 additional peptides (; Table S1), indicating increased production of antibodies targeting these antigens. Some of the peptides above the cut-off level that are not actually encoded within the B. mallei genome were detected by the array. However, as discussed by Waag et al.,Citation10 prior to working at USAMRIID the subject had worked with both B. pseudomallei and B. mallei and thus may have elevated levels of antibodies to some peptides due to previous exposures.
Table 1. Highly increased antibodies reactivity in a human glanders infection
Antibodies against five different type III secretion system components were highly increased in the human glanders infection (), four of which (BPSS1390/BMAA1602, BPSS1401/BPSS1620/BMAA1630 and BPSS1534/BMAA1532) were not seen in melioidosis serum from patients in southeast Asia.Citation11 BPSS1532/BMAA1530 was seen in the glanders infection as well as in melioidosis patients and healthy controls from southeast Asia (). The type III secretion system is a bacterial protein export mechanism that forms syringe-like appendages present in several bacterial species that function to inject effector molecules into host cells. The type III secretion system has been shown to be required for virulence in mouse and hamster models for B. mallei and B. pseudomallei (reviewed by Galyov et al.Citation5).
Table 2. Comparison of the antibody profiles of serum from human glanders, recovered melioidosis patients and healthy controls from southeast Asia
Red, human glanders; blue, recovered melioidosis patients;Citation11 yellow, healthy controls from southeast Asia.Citation11 ORFs in the accompanying list are color coded to match the Venn diagram. NA, gene not present in B. mallei.
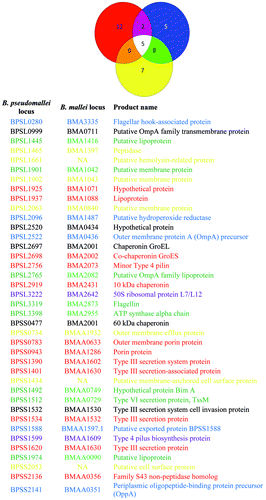
Type 4 pili are complex structures used by Gram-negative and Gram-positive bacteria for motility and surface attachment.Citation14 The B. mallei major pilin, PilA, and B. pseudomallei minor pilin, PilV, have both been shown to be immunogenic, but failed to protect mice against challenge.Citation15,Citation16 We noted antibodies against PilA (BPSL0782/BMA0278) were increased slightly in the serum from the glanders infection (Table S1), while antibodies against a different minor pilin of the Type 4 pili system (BPSL2756/BMA2073) were increased 2-fold (); PilV (BPSS1593) was not represented on the array. Antibodies against the Type 4 pilus component BPSS1599/BMAA1609 were detected in human glanders serum and serum from recovered melioidosis patientsCitation11 ().
In addition to antibodies to recognized virulence factors, antibodies against a lipoprotein (BPSS1937/BMA1088), three porins/outer-membrane proteins (BPSL0999/BMA0711, BPSS0943/BMAA1286 and BPSS0783/BMAA0633) and four chaperonins (BPSL2697/BPSS0477/BMA2001, BPSL2919/BMA2431 and BPSL2698/BMA2002) were strongly increased above background (). While little is known about the role of lipoproteins in B. mallei and B. pseudomallei pathogenesis, two lipoproteins not represented on this array have been identified in previous B. pseudomallei studies: a signature-tagged mutagenesis experiment identified BPSL3147 (BMA2723) as being required for virulence in mice,Citation17 while immunization with BPSL2151 (BMA1547) was shown to provide protection from, but not clearance of, B. pseudomallei in mice.Citation18 Porins and outer-membrane proteins have been characterized in membrane preparations of B. mallei and B. pseudomallei.Citation19 However, only one of the porins (BPSS2136/BMAA0356) that reacted at elevated levels with the human glanders serum () was detected as one of the top 20 proteins in B. mallei outer membrane preparations.Citation19 A second protein, BPSS0943/BMAA1286, was also detected in the outer membrane preparationsCitation19 and highly elevated in the human glanders infection (). These data suggest that not all outer membrane proteins in B. mallei are equally antigenic.
Using this serum, Amemiya and colleagues previously identified via ELISA that IgG against GroEL increased ~10-fold and anti-DnaK IgG increased ~1.5-fold.Citation9 The present study showed that antibodies against GroEL (BPSL2697/BMA2001) were increased ~4.8-fold while antibodies against DnaK (BPSL2827/BMA2326) were increased ~1.4-fold (; Table S1). The difference in observed levels of antibodies against GroEL may reflect sensitivity differences between the technologies.
Due to limited data available for B. mallei infections, it is impossible to evaluate these results in the context of existing literature without the obvious comparisons to the genetically related B. pseudomallei. The distinctive antibody profile from this glanders infection compared with existing melioidosis literature suggests some interesting contrasts between the pathogenesis of these two diseases. A recent study used the same protein array platform to probe antibody response in individuals who had recovered from melioidosis in southeast Asia.Citation11 We noted some overlap between the antibodies identified in recovered patients and healthy controls and those from recovered patients with the results from the human glanders infection ( and ). Interestingly, there was only minor overlap between the antibody reactivity found in the glanders serum and that from the melioidosis patients. Only antibodies against BPSS1599 (Type 4 pilus biosynthesis protein) and BPSL3222 (50S ribosomal protein L7/L12) were elevated in both infections and not present in serum from healthy humans in southeast Asia ( and ).
Differences were noted in antibodies produced from this human infection and reports on antibodies from horses with glanders. Using phage display technology, Tiyawisutsri et al. screened equine glanders infection serum and identified antibodies against four chromosomal loci that were over-represented in their library.Citation20 Two of these four loci encoded a total of three peptides present on the array (BMAA1324, BMA1024 and BMA1027), but none of them had greatly increased antibodies in the human infection (Table S1). The reason for these differences is not known, but it could be due to the different screening technology, the fact that horses are prone to a chronic glanders infection while the human case was acute and/or different immunogenic antigens that are prominent in these different hosts.Citation5,Citation8
These data can also be compared with the outer membrane proteome of B. mallei.Citation19 The general absence of proteins identified by screening for the presence of, and increase in, antibodies compared with the proteome dataCitation19 is intriguing as it would be anticipated that highly expressed proteins would overlap with the proteins that elicited the strongest antibody response. These data suggest that while proteins may be highly expressed in vitro, they are either not highly expressed in vivo or may be non-immunogenic.
As this approach has shown promise and greatly expands on existing research, it warrants further studies using an animal model so that proper statistical analyses and comparisons may be performed. This will also allow for a comparison between host data in order to verify that antibodies produced in mouse infections are representative of antibodies produced in a human infection. In other studies protection in mice can be achieved with monoclonal antibodies against B. mallei administered prior to, but not after, challenge.Citation21 However, in these studies the animals’ spleens were heavily colonized with B. mallei despite surviving the infectionCitation21 and a similar result was observed with a lipoprotein vaccination of B. pseudomallei.Citation18 This current work presents data and identifies potential immunogenic antigens that may be exploited to develop new protective antibodies that overcome this limitation.
As the report by Waag et al. shows,Citation10 serum from this individual reacted to killed whole cells of B. mallei and B. pseudomallei. While that approach allows for serodiagnosis of exposure, it is non-specific. Having a detailed comparison will greatly aid in the development of serodiagnostic antibodies for B. mallei and B. pseudomallei infections. When these human glanders results were compared with serum from recovering melioidosis patients and healthy controls from southeast AsiaCitation11 there were 12 antibodies that were highly increased only in the glanders infection while five were highly present only in the melioidosis samples (). Additionally, seven antibodies were highly present only in the healthy controls while five were detected for all three conditions (). Using a Yersinia pestis protein microarray, Keasey et al. showed that cross reactive antibodies are generated to proteins from number of Gram-negative pathogens, including B. mallei and B. pseudomallei.Citation22 However, the only protein that was cross-reactive and common between the protein microarray used in our study and the Y. pestis protein microarray was GroEL. This result suggests that the 12 proteins which generated antibodies found only in glanders serum represent candidate antigens for the differentiation of glanders and melioidosis infections in humans and that these also have less risk for cross-reactivity with other pathogens.
Additional material
Download Zip (276.5 KB)Acknowledgments
This work was supported by the National Institutes of Health (R21AI73923 to J.B.G., U54AI065359 and U01AI061363 to P.F., and J.J.V. was supported by 5T32AI055432 to the University of Virginia).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/virulence/article/22056
References
- Larsen JC, Johnson NH. Pathogenesis of Burkholderia pseudomallei and Burkholderia mallei. Mil Med 2009; 174:647 - 51; PMID: 19585782
- Wheelis M. First shots fired in biological warfare. Nature 1998; 395:213; http://dx.doi.org/10.1038/26089; PMID: 9751039
- Regis E. The Biology of Doom. New York, NY: Random House, 1999.
- Abilek K, Handleman S. Biohazard: The Chilling True Story of the Largest Covert Biological Weapons Program in the World. New York, NY: Random House, 1999.
- Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol 2010; 64:495 - 517; http://dx.doi.org/10.1146/annurev.micro.112408.134030; PMID: 20528691
- Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, et al. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci U S A 2009; 106:13499 - 504; http://dx.doi.org/10.1073/pnas.0812080106; PMID: 19666533
- Howe C, Miller WR. Human glanders; report of six cases. Ann Intern Med 1947; 26:93 - 115; PMID: 20278465
- Srinivasan A, Kraus CN, DeShazer D, Becker PM, Dick JD, Spacek L, et al. Glanders in a military research microbiologist. N Engl J Med 2001; 345:256 - 8; http://dx.doi.org/10.1056/NEJM200107263450404; PMID: 11474663
- Amemiya K, Meyers JL, Deshazer D, Riggins RN, Halasohoris S, England M, et al. Detection of the host immune response to Burkholderia mallei heat-shock proteins GroEL and DnaK in a glanders patient and infected mice. Diagn Microbiol Infect Dis 2007; 59:137 - 47; http://dx.doi.org/10.1016/j.diagmicrobio.2007.04.017; PMID: 17908615
- Waag DM, England MJ, DeShazer D. Humoral immune responses in a human case of glanders. Clin Vaccine Immunol 2012; 19:814 - 6; http://dx.doi.org/10.1128/CVI.05567-11; PMID: 22398248
- Suwannasaen D, Mahawantung J, Chaowagul W, Limmathurotsakul D, Felgner PL, Davies H, et al. Human immune responses to Burkholderia pseudomallei characterized by protein microarray analysis. J Infect Dis 2011; 203:1002 - 11; http://dx.doi.org/10.1093/infdis/jiq142; PMID: 21300673
- Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 2004; 101:14240 - 5; http://dx.doi.org/10.1073/pnas.0403302101; PMID: 15377794
- Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, et al. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci U S A 2004; 101:14246 - 51; http://dx.doi.org/10.1073/pnas.0403306101; PMID: 15377793
- Varga JJ, Nguyen V, O’Brien DK, Rodgers K, Walker RA, Melville SB. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol Microbiol 2006; 62:680 - 94; http://dx.doi.org/10.1111/j.1365-2958.2006.05414.x; PMID: 16999833
- Fernandes PJ, Guo Q, Waag DM, Donnenberg MS. The type IV pilin of Burkholderia mallei is highly immunogenic but fails to protect against lethal aerosol challenge in a murine model. Infect Immun 2007; 75:3027 - 32; http://dx.doi.org/10.1128/IAI.00150-07; PMID: 17403869
- Sangdee K, Waropastrakul S, Wongratanachewin S, Homchampa P. Heterologously type IV pilus expressed protein of Burkholderia pseudomallei is immunogenic but fails to induce protective immunity in mice. Southeast Asian J Trop Med Public Health 2011; 42:1190 - 6; PMID: 22299445
- Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PC, Titball RW, et al. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun 2007; 75:1186 - 95; http://dx.doi.org/10.1128/IAI.01240-06; PMID: 17189432
- Su YC, Wan KL, Mohamed R, Nathan S. Immunization with the recombinant Burkholderia pseudomallei outer membrane protein Omp85 induces protective immunity in mice. Vaccine 2010; 28:5005 - 11; http://dx.doi.org/10.1016/j.vaccine.2010.05.022; PMID: 20546831
- Schell MA, Zhao P, Wells L. Outer membrane proteome of Burkholderia pseudomallei and Burkholderia mallei from diverse growth conditions. J Proteome Res 2011; 10:2417 - 24; http://dx.doi.org/10.1021/pr1012398; PMID: 21391724
- Tiyawisutsri R, Holden MT, Tumapa S, Rengpipat S, Clarke SR, Foster SJ, et al. Burkholderia Hep_Hag autotransporter (BuHA) proteins elicit a strong antibody response during experimental glanders but not human melioidosis. BMC Microbiol 2007; 7:19; http://dx.doi.org/10.1186/1471-2180-7-19; PMID: 17362501
- Treviño SR, Permenter AR, England MJ, Parthasarathy N, Gibbs PH, Waag DM, et al. Monoclonal antibodies passively protect BALB/c mice against Burkholderia mallei aerosol challenge. Infect Immun 2006; 74:1958 - 61; http://dx.doi.org/10.1128/IAI.74.3.1958-1961.2006; PMID: 16495574
- Keasey SL, Schmid KE, Lee MS, Meegan J, Tomas P, Minto M, et al. Extensive antibody cross-reactivity among infectious gram-negative bacteria revealed by proteome microarray analysis. Mol Cell Proteomics 2009; 8:924 - 35; http://dx.doi.org/10.1074/mcp.M800213-MCP200; PMID: 19112181