Abstract
Candida albicans is a major fungal pathogen of humans, causing mucosal infections that are difficult to eliminate and systemic infections that are often lethal primarily due to defects in the host’s innate status. Here we demonstrate the utility of Caenorhabditis elegans, a model host to study innate immunity, by exploring the role of reactive oxygen species (ROS) as a critical innate response against C. albicans infections. Much like a human host, the nematode’s innate immune response is activated to produce ROS in response to fungal infection. We use the C. albicans cap1 mutant, which is susceptible to ROS, as a tool to dissect this physiological innate immune response and show that cap1 mutants fail to cause disease and death, except in bli-3 mutant worms that are unable to produce ROS because of a defective NADPH oxidase. We further validate the ROS-mediated host defense mechanism in mammalian phagocytes by demonstrating that chemical inhibition of the NADPH oxidase in cultured macrophages enables the otherwise susceptible cap1 mutant to resists ROS-mediated phagolysis. Loss of CAP1 confers minimal attenuation of virulence in a disseminated mouse model, suggesting that CAP1-independent mechanisms contribute to pathogen survival in vivo. Our findings underscore a central theme in the process of infection—the intricate balance between the virulence strategies employed by C. albicans and the host’s innate immune system and validates C. elegans as a simple model host to dissect this balance at the molecular level.
Keywords: :
Introduction
The incidence of invasive fungal infections has escalated in recent years, primarily in hospital settings;Citation1 90% of these infections are caused by various Candida species,Citation2 50% of which are fatal.Citation3 The estimated annual cost of treating nosocomial Candida infections exceeds $1 billion per year and has an attributable mortality of about 5,000 deaths per year in the United States.Citation4-Citation6 Candida species are the fourth leading cause of bloodstream infections (BSI), with C. albicans responsible for about half the cases. C. albicans form robust biofilms on medical implants, such as intravenous catheters, prosthetic joints, or artificial heart valves, which can seed potentially lethal disseminated infections.Citation7-Citation9 Approximately 75% of women have at least one episode of vaginitis caused by Candida in their lifetimeCitation10 and oropharyngeal thrush and esophagitis are common in both infants and in patients with AIDS.Citation11 Candida species also cause superficial infections on mucosal surfaces in the body, including the mouth, upper gastrointestinal (GI) and urogenital tract. The frequency of these superficial infections combined with the treatment challenges posed by disseminated infections make C. albicans an important pathogen for further study.
A variety of in vitro, ex vivo and in vivo models have been employed to study the interaction between the host and this fungal pathogen. In vitro studies of hyphal morphogenesis and biofilm formation, among many others, have yielded important insights into virulence.Citation12,Citation13 Ex vivo models, such as co-culturing C. albicans with isolated macrophages, neutrophils, epithelial or endothelial cells, and even intact, perfused organs, have demonstrated that C. albicans has very complex responses to host cell contact, which can differ dramatically between cell types.Citation14-Citation19 A murine model of disseminated candidiasis has been frequently used to validate the role of specific genes on overall virulence. There is a general appreciation that each of these models has provided important insights into fungal pathogenesis. Recently, invertebrate models have become additional tools to dissect the roles of components of the antifungal host defense system, including flies (Drosophila melanogaster), wax moth larvae (Galleria mellonella) and the nematode Caenorhabditis elegans.Citation20-Citation22
C. elegans has emerged as a useful model to study infectious disease for several reasons. First, facets of its innate immune system are conserved in humansCitation23,Citation24 and the nematode reacts to pathogens in a manner similar to mammals, such as activation of specific signal transduction pathways.Citation23-Citation26 A rich body of literature demonstrates that human pathogens, both bacteria and fungi, also infect C. elegans in ways that are mechanistically similar to humans. For example the opportunistic human pathogens Pseudomonas aeruginosaCitation23,Citation27-Citation30 and Serratia marcescensCitation24,Citation31 produce toxins that are required for pathogenesis in disparate eukaryotic hosts. Mutant studies in Salmonella Typhimurium, typically thought to have a narrow host range, shows a direct correlation in virulence between humans and C. elegans.Citation32,Citation33 More recently a comparative study in C. elegans using fungal pathogens of the genus CryptococcusCitation34,Citation35 showed that only the human pathogen C. neoformans, but not other related yeasts (C. kurtzingii or C. laurentii), killed the nematode. These studies further demonstrated that a virulence factor such as Kin1, first identified in nematodes, was also important in mammals.Citation36 More recent whole genome analyses of C. elegans infected with C. albicans reveal that the nematode induces immune defenses with known antifungal properties.Citation26
Studies of C. elegans infected with bacterial pathogens reveal that the generation of reactive oxygen species (ROS) is an important part of the nematode’s defense response,Citation25,Citation37 a hallmark shared with mammalian innate immune responses. We previously reported a C. elegans-based assay to study several aspects of disease progression, namely Dar (deformity in the anal region), an early marker of infection; intestinal distension, resulting from colonization of the intestine; swelling in the vulva, representing infection of other epithelial layers; and ultimately death of the host worm.Citation25 This assay mimics an infection, because a small amount of the pathogen is introduced along with the food. Furthermore the assay is amenable to unbiased, high-throughput screens because it does not require manual handling of individual animals. Developed initially using Saccharomyces cerevisiae, here we have adapted the assay for C. albicans.
We use genetic and pharmacological tools to alter the intricate balance between the host and the pathogen and demonstrate that ablating either the ability of the worm to produce ROS or C. albicans to detoxify it (via mutation of the Cap1 transcription factor) has dramatic effects on the outcome of this infection. Results in the worm were recapitulated in macrophage co-cultures, validating this model. Surprisingly, the C. albicans cap1Δ mutant retained limited virulence in the disseminated murine bloodstream model, suggesting that additional layers of regulation of antioxidant defense exist in the context of a mammal. This work thus provides an avenue to investigate fungal pathogenesis and has allowed us to identify further complexity in the pathogenic C. albicans-host interaction.
Results
Generation of CAP1 homozygous and complemented strains
The original cap1Δ/Δ mutant strain CJD21 was generated by Raymond and colleaguesCitation38 using an approach that was subsequently shown to be inappropriate for animal experiments due to the variability of expression of the URA3 selectable marker. We selected a ura3-derivative of the cap1Δ/Δ strain CJD21 on 5-FOA, then integrated either URA3 or URA3-CAP1 at the RPS10 locus using plasmid CIp10, a strategy shown to stably express URA3 during infection.Citation39 This generated a cap1Δ/Δ mutant strain (AGC2) and a complemented strain (AGC4); the mutant strain is phenotypically identical to the original CJD21 during in vitro challenge with ROS (data not shown).
In vivo virulence assay for Candida infection
Here we report a pathogenesis assay to test various fungal pathogens of the Candida genus, including C. albicans, C. dubliniensis, C. krusei, C. tropicalis and C. glabrata. We adapted a previously described pathogenesis assay where a small quantity of S. cerevisiae, was introduced with the nematode’s standard diet of E. coli OP50. E. coli was attenuated to avoid interactions with the fungal species under investigation.Citation25 We decreased the concentration of Candida in the feeding mixture by 30-fold because of the increased pathogenicity of Candida species, and even at this lower fungal burden, most species induced Dar in 100% worms, compared with S. cerevisiae, which induced 0% Dar at the same concentration of inoculum (). We uncovered molecular mechanisms of fungal virulence and the reciprocal innate immune response of nematodes, which is also conserved in mammals.Citation40,Citation41
Table 1. Percentage Dar observed for different clinical isolates
Disease phenotypes of C. elegans upon C. albicans infection
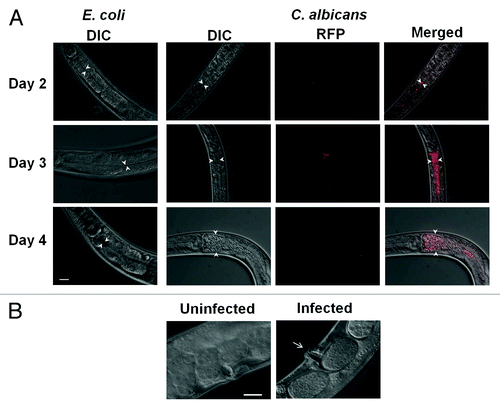
Subsequent studies focused on C. albicans because it is the most prevalent infectious species in this genusCitation3 and adequate genomic and molecular tools have been developed. To visualize infection, disease progression and death, we exposed nematodes to C. albicans and observed them daily over a 6 d period. The Dar phenotype was clearly visible in every worm on Day 4 ( and ). Swelling in the vulvar region was also noted in the worms infected with C. albicans (). The worm succumbed to the infection following these phenotypic observations. To observe colonization of the intestinal lumen, worms were exposed to mCherry-labeled C. albicans (mCherry labeled SC5314, gift from Dr. Robert Wheeler, Univ. of Maine). Time-series micrographs of worms infected with C. albicans indicated that the intestinal lumen was colonized 2 d post-exposure compared with uninfected worms. Considerable intestinal distension was observed in infected worms on subsequent days () compared with the uninfected control population. In general, the disease phenotypes were more robust with C. albicans compared with S. cerevisiae, even with lower inoculum for infection. To further validate our assay we tested known mutants efg1Δ/Δ and cph1Δ/Δ—previously documented virulence factors that regulate hyphal transition of C. albicansCitation42 and have been shown to be important in in vivo infections of mice and nematodes.Citation26,Citation43 The efg1Δ/Δ and cph1Δ/Δ single mutants showed decreased Dar 10% and 50%, respectively, compared with the cognate wild type, while the efg1Δ/Δ cph1Δ/Δ double mutant failed to induce any Dar (0% Dar). These results recapitulate the pattern of virulence in mice, where the cph1Δ/Δ mutant is slightly attenuated, the efg1Δ/Δ mutant is significantly attenuated, and the double mutant is completely avirulent.Citation42 Accordingly, our assay is suitable for exploring virulence strategies of C. albicans and the reciprocal host defenses that may be correlated with aspects of innate immunity that is conserved in mammals.Citation23,Citation24
Figure 1.C. albicans induces deformed anal region (Dar) in wild-type worms. Worms were exposed to E. coli as control (uninfected) and C. albicans (shown are two examples of infected worms) and pictures were taken on Day 4 (arrow indicates the Dar region). Scale = 20 µm.
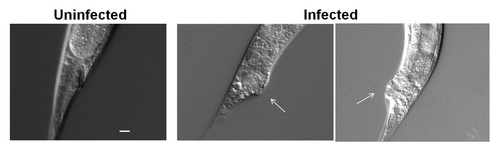
Figure 2. Different phenotypes are observed due to Candida infection. (A) Exposure to C. albicans causes intestinal distention in the worms over time (Days 2, 3 and 4). DIC (Nomarski) pictures of worms feeding on E. coli was taken as control and the DIC, RFP and merged pictures of worms infected with C. albicans are shown over 3 d. (B) Vulva swelling is observed (arrow points to the vulvar region) by Day 4 when worms are exposed to Candida compared with worms exposed E. coli as control (uninfected). Scale = 20 µm.
CAP1 is required to establish and sustain infection in nematodes
We compared the Dar response of worms exposed to null mutant (cap1Δ/Δ) and wild-type strain (CAP1/CAP1) as well as a CAP1-complemented strain (cap1Δ/Δ + CAP1), where a single wild-type copy of CAP1 has been re-introduced. The wild-type strain was able to induce Dar in 100% of the worms while the cap1Δ/Δ homozygous mutant showed a significant reduction in Dar induction (). The CAP1-complemented strain showed an intermediate Dar response that is commonly observed for complemented strains. This result indicates that Cap1 function is required for full virulence in the nematode model.
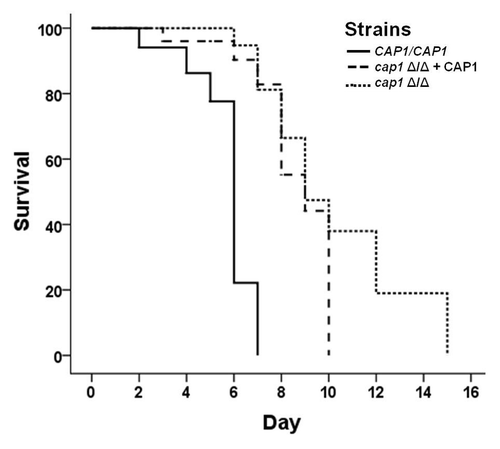
Figure 3. Cap1 is a virulence factor that is required for counteracting oxidative stress created by the host during fungal pathogenesis. The Dar phenotype is observed for both (A) wild-type and (B) bli-3 mutant worms (which lack the ability to produce ROS) exposed to CAP1/CAP1, cap1Δ/Δ + CAP1 and cap1Δ/Δ strains (* denotes p < 0.001; n denotes the number of worms exposed to a particular strain). Experiment was done in triplicate. (C) Survival curves for wild-type worms when exposed to CAP1/CAP1, cap1 Δ/Δ + CAP1 and cap1 Δ/Δ with E. coli OP50 as control (p < 0.01 for CAP1/CAP1 and the cap1Δ/Δ mutant) show that worms exposed to cap1 Δ/Δ survive longer than worms exposed to wild type, while (D) bli-3 mutant worms exposed to Candida strains show no difference in survival between exposure to wild-type CAP1/CAP1 and the cap1Δ/Δ mutant.
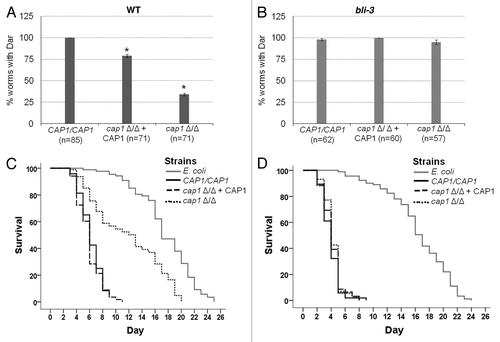
To test whether the mechanism by which Cap1 promotes virulence is related to the production of ROS by the host, we tested mutants in the bli-3 gene which contains the only NADPH oxidase moiety in C. elegans genome.Citation47 We and others previously reported that Bli-3 produces ROS in response to pathogenic insult.Citation25,Citation37 To test the hypothesis that Cap1 is responsible for counteracting this oxidative environment upon C. albicans infection, we exposed bli-3 mutant worms to cap1Δ/Δ, CAP1-complemented and wild-type CAP1/CAP1 strains. As shown in , the Dar phenotype of bli-3 mutant worms infected with the cap1Δ/Δ null mutant is indistinguishable from that of the wild type and complemented counterparts, suggesting that Cap1 function is not required for infection if the host cannot produce ROS. Thus, Cap1 is dispensable when the ROS production part of the host’s defense repertoire has been compromised, as in the bli-3 mutant.
We and others previously showed that Dar is an early indicatorCitation25,Citation48 of an eventually lethal infection. To test the hypothesis that cap1Δ/Δ mutants are avirulent, we measured the life span of worms infected with the cap1Δ/Δ null mutant, complemented and wild-type strains relative to uninfected worms. The average lifespan of C. elegans infected with wild-type C. albicans (CAP1/CAP1) or a CAP1-complemented strain was significantly shorter (median survival = 6 d) than those infected with the cap1Δ/Δ null mutant (median survival = 13 d) (). These data indicate that Cap1 is required for the infection to persist and ultimately kill the host. To test the hypothesis that Cap1 is required to neutralize host ROS, we challenged an ROS-deficient bli-3 mutant host with the C. albicans cap1Δ/Δ mutant. Lifespan plots reveal that the kinetics of killing are indistinguishable between the cap1Δ/Δ mutant and the wild type or complemented strains (), suggesting that ROS is the primary defense of nematodes against C. albicans and that C. albicans requires Cap1 for a lethal infection to persist. The lifespan of uninfected bli-3 mutants is the same as their wild-type counterparts (), eliminating confounding factors other than ROS production as a likely cause of death of infected worms.
In vitro, Cap1 regulates the response to oxidative stress,Citation38,Citation44-Citation46 which is a key component of the human innate immune response to fungal infections by neutrophils.Citation15 To correlate these studies, performed at 30°C and 37°C, with our in vivo infection, performed at 20°C, we compared the ROS sensitivity profiles of cap1Δ/Δ, CAP1-complemented and the cognate wild-type strain at all three temperatures using hydrogen peroxide. The results indicate that ROS sensitivity of the cap1Δ/Δ strain is independent of temperature and that all C. albicans strains used in this study show the same phenotype at all temperatures (data not shown).
Cap1 is responsible for neutralizing ROS produced by phagocytes
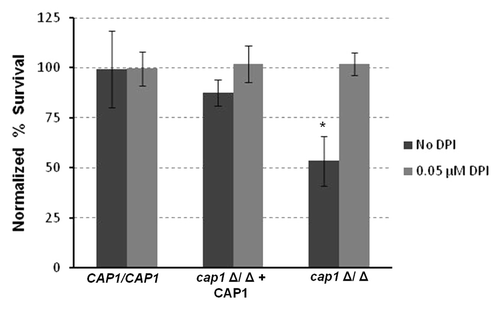
Part of the mammalian host defense against Candida infection is production of ROS within phagosomes that have engulfed the fungal pathogen.Citation49 Wild-type C. albicans can effectively neutralize ROS to surviveCitation14,Citation50-Citation53 and eventually cause macrophages to lyse. To test whether Cap1 is important in surviving phagocytosis, we co-cultured macrophages with either wild type or cap1Δ/Δ strains. Our data indicates that cap1Δ/Δ strains are significantly more likely to be killed by macrophages than the wild-type or complemented strains (). This effect was eliminated when the co-cultures were treated with diphenyleneiodonium chloride (DPI), an inhibitor of the phagocyte oxidase that generates ROS, thus demonstrating that host-derived ROS kills C. albicans (). These results mimic our genetic studies in nematodes and suggest that C. albicans relies on Cap1 to neutralize the oxidative environment within the phagosome, and that eliminating the source of ROS in phagosomes eliminates the need for Cap1.
Figure 4. Cap1 is required for survival in macrophages. Macrophages were exposed to different strains of Candida in a ratio of 1:15 macrophages with and without DPI. Percentage survival was calculated by dividing the CFUs obtained for Candida grown with macrophages to Candida alone. Data were then normalized to the wild type. Percent survival of cap1Δ/Δ mutant is significantly less (**p < 0.05) than the wild type and the complemented Cap1 strain.
CAP1 has a limited effect during mammalian infection
The cap1Δ/Δ mutant is clearly attenuated in the nematode model and during contact with macrophage, and has previously been reported to be hypersensitive to several sources of oxidative stress.Citation38 We tested it in the standard mouse model of disseminated candidiasis, in which C. albicans is introduced directly into the circulatory system via the tail vein. We injected outbred ICR mice with 106 C. albicans cells and monitored for signs of infection (; see Materials and Methods). Mice inoculated with the cap1Δ/Δ strain had a statistically significant increase in median survival times relative to wild type (from 6.0 to 8.5 d, p = 0.0003). However, the CAP1-complemented strain showed roughly the same virulence (median survival 8.0 d). A second set of cap1Δ/Δ strains, generated using the SAT1-flipper methodCitation54 also showed little difference from the wild-type in the mouse model (data not shown). Thus, despite the worm and macrophage data, in vitro cap1Δ/Δ phenotypes and the general assumption that the oxidative burst is a key part of the anti-fungal innate immune response, we show that loss of CAP1 alters virulence only slightly.
Figure 5. Candida virulence is reduced when Cap1 is deleted. Ten mice/strain were injected in the tail vein with 106 cells of the respective Candida strain. Survival curves of mice when infected with CAP1/CAP1, cap1Δ/Δ + CAP1 and cap1 Δ/Δ strains show that mice infected with the cap1Δ/Δ mutant were able to survive significantly longer than those injected with the wild type (p < 0.01 for CAP1/CAP1 and the cap1Δ/Δ mutant).
Discussion
Mammalian biology has been effectively modeled using a variety of species that present substantial advantages in complexity, genetic tractability, ethical considerations and cost. The last decade has seen the acceptance of invertebrates, such as C. elegans, as relevant hosts that can contribute to the molecular understanding of microbial pathogenesis. C. elegans has proven an effective model for several systemic pathogens such as Cryptococcus, Pseudomonas and Enterococcus, among others,Citation32,Citation55 even though it differs from mammalian infections in several respects, including the site of infection (gut vs. bloodstream), growth temperature (25–30°C vs. 37°C) and the absence of an adaptive immune response. We previously demonstrated that the model yeast S. cerevisiae can cause pathology in the nematode.Citation25 In this work we show that this assay is far more robust when using pathogenic Candida species and that the virulence of these species in the worm roughly correlates with virulence in humans.
A common feature of the innate immune response across metazoans is the generation of ROS via an NADPH-dependent oxidase, the phagocyte oxidase (a complex of gp47phox and gp91phox) in mammals and bli-3 in the worm.Citation37,Citation56,Citation57 In humans, chronic granulomatous disease is caused by mutations in genes encoding different subunits of NADPH-oxidase and 60% of these patients, show loss of gp91phox expression.Citation58 Interestingly these patients are more susceptible to fungal infections mainly Aspergillus and Candida and have a high mortality rate.Citation59 Mice lacking the gp91phox subunit are more susceptible to disseminated infection with C. albicansCitation60 and knocking down gp47phox in zebrafish increases the severity of infection.Citation61 We show here that host-generated ROS is also protective against C. albicans infection in the invertebrate C. elegans. Moreover, Cap1, a fungal-specific transcription factor that regulates oxidative stress responsesCitation38,Citation45 and drug resistance pumpsCitation62 in vitro is required for virulence of C. albicans in nematodes and survival in cultured macrophages. Specifically, strains lacking CAP1 induce Dar phenotype less frequently and attenuate virulence in a worm-killing assay relative to the wild-type strain. This is a direct consequence of increased sensitivity of this mutant to ROS. Worms that cannot produce ROS due to a mutation in the host oxidase show early signs of disease, and succumb to an infection with the cap1Δ/Δ null mutant. Furthermore when the NADPH oxidase is chemically inactivated in cultured macrophages, cap1Δ/Δ null mutants are able to survive just as well as their wild-type counterpart.
Given the data above and the known in vitro functions of CAP1 in responding to ROS, the results of the murine model are surprising, as we cannot conclude that loss of CAP1 alters virulence. Since some other antioxidant proteins, such as the secreted superoxide dismutase SOD5 and catalase CAT1, are attenuated in this same model,Citation36,Citation53,Citation63 it seems clear that anti-oxidant defenses are important in vivo. A more intriguing possibility is that there may be Cap1-independent mechanisms for their induction in the context of the mammalian host. Indeed, the superoxide dismutases SOD4 and SOD5 are both more highly expressed in hyphal cells, which can be induced in response to several host-associated cues.Citation51,Citation63 Thus, our C. elegans model may allow analysis of critical fungal responses to the innate immune system that could be masked by phenotypic redundancy in the context of the intact mammal. In conclusion, host-generated ROS is a critical component of the ancient innate immune response against C. albicans in a variety of species and proper regulation of anti-oxidant defenses is an important virulence factor in many species including, as we show here, in C. albicans.
Materials and Methods
Strains, media and growth conditions
The C. albicans strains used are listed in and are based on SC5314 and its auxotrophic derivative CAI4-F2. C. albicans transformations were performed via electroporation.Citation54 Fungal growth medium was prepared as described previouslyCitation64 and strains were grown overnight in yeast extract-peptone-dextrose (YPD) at 37°C. The C. elegans strains were grown at 20°C on nematode growth agar medium (NGM), spotted with Escherichia coli OP50 and maintained as described previously.Citation65 E. coli OP50 was grown overnight in Luria broth at 37°C.
Table 2. Strains used in this study
Generation of cap1 mutant strains
The existing cap1Δ/Δ strain CJD21Citation38 expresses URA3 from the disrupted cap1 locus, a strategy that has been subsequently demonstrated to potentially affect virulence through mis-expression of the URA3 marker;Citation66,Citation67 this can be overcome through ectopic integration of URA3 at the RPS10 locus using plasmid CIp10.Citation39,Citation68 To generate the cap1Δ/Δ mutant and complemented strains with URA3 at RPS10, we grew CJD21 on YPD for two overnight passages in YPD, then plated to media containing 5-fluororotic acid (5-FOA) to select for ura3 auxotrophs.Citation69 PCR amplified the CAP1 open reading frame from genomic DNA of strain SC5314, plus ~1,000 bp of 5′ UTR and ~350 bp of 3′ UTR. This fragment was ligated between the XhoI and HinDIII sites in CIp10. This plasmid was digested with StuI and used to transform the 5-FOA-selected cap1Δ/Δ strain. In parallel, the empty CIp10 plasmid was digested with StuI and used to transform the same strain. After selection on SD-Ura, correct integration at RPS10 was confirmed by PCR to generate cap1Δ/Δ strains with either URA3 or CAP1–URA3 expressed from the same genomic site as the mutant (AGC2) and complemented (AGC4) strains, respectively.
Microscopic analysis of C. elegans
A 2% agarose pad containing 0.01 M sodium azide as anesthetic was prepared on a slide. A 5 µl drop of M9 buffer was added to the pad. Worms exposed to wild-type Candida or RFP-labeled Candida were picked and transferred to the drop on the slide. Mounted worms were then covered with a coverslip and observed at 200× and 400× magnifications using an Axiovision Zeiss microscope under differential interference contrast (Nomarski) and epifluorescence optics. An ApoTome attachment was used to enhance the fluorescence images.
Egg preparation
Three worms each in the L3/L4 stage were transferred to two NGM agar plates containing E. coli OP50 and grown at 20°C for 4 d. On the day of the experiment, worms were washed off the plates with M9 buffer and centrifuged at 900 × g for 2 min. The supernatant was removed and the worms were then re-suspended in a bleach solution [1:4 dilution of commercial bleach (5.25%) containing 0.25 M sodium hydroxide]. The worm suspension was mixed gently by inversion for 3 min and centrifuged for 2 min at 2,000 × g. The pellet was washed and centrifuged with M9 buffer at 2,000 × g for 2 min and then finally re-suspended in 500 µl M9 buffer. The egg suspension was diluted or concentrated with M9 buffer as required to obtain approximately 30–40 eggs/5 µl.
Pathogenesis assay
E. coli and Candida strains were grown overnight at 37°C. Culture aliquots were centrifuged at full speed for 1 min in a table top microcentrifuge and the supernatant removed. Pellets were washed twice in sterile deionized water, and re-suspended to a final concentration of 200 mg/ml and 10 mg/ml, respectively. Next, a mixture of 10 µl of a 50 mg/ml streptomycin sulfate stock to inhibit E. coli growth, 7 µl of distilled water, 2.5 µl of E. coli and 0.5 µl of Candida was spotted on to each NGM plate. E. coli spotted plates were used as control. Finally, 5 µl of C. elegans egg suspension was transferred to each plate. Plates were then kept in a 20°C incubator and were observed for 5 d. All the experiments were done in triplicate. Student’s t-test was used to check the statistical significance of the differences observed between wild type and other Candida strains.
C. elegans survival analysis
Survival analysis was done on the CAP1/CAP1, cap1Δ/Δ + CAP1 and cap1Δ/Δ strains and plates were prepared as described under egg preparation and pathogenesis assay. Starting on Day 3 following the initial assay, the number of worms alive, dead and dead on the rim was recorded each day. In order to differentiate between the first-generation worms and their offspring, worms were transferred every two days to new plates that were prepared the day before as previously discussed and incubated overnight at 20°C.
For data analysis, SPSS (IBM, Inc.) was used to generate Kaplan-Meier survival curves. In this experiment, each worm that died on the plate was entered as a “1,” indicating the event of death due to fungal disease took place. Worms that were found dead on the rim of the plate were censored and entered as a “0,” since death occurred for a non-related reason. Significance, as defined as a p value < 0.05, was assessed using the Gehan-Breslow test. This test assumes that data from earlier survival times are more accurate than later times and weights these data accordingly. Data were combined from three plates and another independent experiment gave the same results.
Macrophage growth inhibition assay
Macrophage cell line RAW 264.7 (ATCC) was used in the assay. The cell line was maintained in DMEM supplemented with 10% fetal bovine serum (FBS). The protocol was slightly modified from the original.Citation70 Briefly, macrophages, on reaching 80–90% confluence, were scraped and brought up in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin, then 2 × 106 macrophage cells were plated on 35 mm2 plates and allowed to adhere for 5 h. Candida strains were grown overnight at 37°C and diluted 1:10 and allowed to grow for another 5 h. Candida cells were then washed with supplemented DMEM and were added to the plates containing macrophages in a ratio 1:15 macrophage and to a final volume of 2 ml. The yeast strain was grown in parallel without macrophages to calculate percent survival. Plates were incubated overnight at 37°C and 5% CO2. Cells were then brought up to 24 ml in a tube using 0.05% Triton X-100 (v/v) in water to osmotically lyse the macrophage cells. Dilutions were prepared and plated on YPD plates and grown overnight at 37°C. Colony forming units (CFUs) were counted and percent survival was calculated by taking the ratio of CFU from co-culture of Candida and macrophage to the CFU obtained for Candida alone.
Experiments involving diphenyleneiodium chloride (DPI) were performed as mentioned above but with the addition of 0.05 µM DPI from a 31.8 mM stock solution in DMSO. Controls without DPI had the same concentration of DMSO. Statistical analysis was done using Student’s t-test.
Mouse virulence assay
Mouse virulence assays were performed as described previously.Citation71 Female, adult (21–25 g) ICR mice (Harlan) were maintained on a normal laboratory diet. C. albicans strains were passaged twice in overnight cultures in YPD, then diluted into fresh YPD and grown for 3 h at 30°C. Cell were collected by centrifugation, washed with water and re-suspended in PBS, then diluted and counted with a hemocytometer. Cells were diluted to 1 × 108 cells/ml in PBS. Mice were injected with 100 µl of this suspension via the tail vein, with groups of 10 mice/strain. Animals were monitored 2–3 times daily for signs of infection and were euthanized when moribund. Survival data were analyzed with Prism5 (Graphpad Software) using the log rank test. All animal experiments were conducted in accordance with protocols approved by the University of Texas Health Science Center Animal Welfare Committee.
Acknowledgments
We would like to thank Dr. Ryder and L. Gaffney for their critical review of the manuscript. This work was supported in part by Public Health Service award R01AI075091 to M.C.L.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Pfaller MA. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin Infect Dis 1996; 22:Suppl 2 S89 - 94; http://dx.doi.org/10.1093/clinids/22.Supplement_2.S89; PMID: 8722834
- Fridkin SK, Jarvis WR. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev 1996; 9:499 - 511; PMID: 8894349
- Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, et al. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis 2003; 37:1172 - 7; http://dx.doi.org/10.1086/378745; PMID: 14557960
- Miller LG, Hajjeh RA, Edwards JE Jr. Estimating the cost of nosocomial candidemia in the united states. Clin Infect Dis 2001; 32:1110; http://dx.doi.org/10.1086/319613; PMID: 11264044
- Pappas PG, Rex JH, Lee J, Hamill RJ, Larsen RA, Powderly W, et al, NIAID Mycoses Study Group. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis 2003; 37:634 - 43; http://dx.doi.org/10.1086/376906; PMID: 12942393
- Morgan J, Meltzer MI, Plikaytis BD, Sofair AN, Huie-White S, Wilcox S, et al. Excess mortality, hospital stay, and cost due to candidemia: a case-control study using data from population-based candidemia surveillance. Infect Control Hosp Epidemiol 2005; 26:540 - 7; http://dx.doi.org/10.1086/502581; PMID: 16018429
- Velasco E, Thuler LC, Martins CA, Nucci M, Dias LM, Gonçalves VM. Epidemiology of bloodstream infections at a cancer center. Sao Paulo Med J 2000; 118:131 - 8; http://dx.doi.org/10.1590/S1516-31802000000500004; PMID: 11018846
- Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect Control Hosp Epidemiol 2000; 21:510 - 5; http://dx.doi.org/10.1086/501795; PMID: 10968716
- Wisplinghoff H, Seifert H, Tallent SM, Bischoff T, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in pediatric patients in United States hospitals: epidemiology, clinical features and susceptibilities. Pediatr Infect Dis J 2003; 22:686 - 91; http://dx.doi.org/10.1097/01.inf.0000078159.53132.40; PMID: 12913767
- Sobel JD. Pathogenesis and epidemiology of vulvovaginal candidiasis. Ann N Y Acad Sci 1988; 544:547 - 57; http://dx.doi.org/10.1111/j.1749-6632.1988.tb40450.x; PMID: 3063184
- Darouiche RO. Oropharyngeal and esophageal candidiasis in immunocompromised patients: treatment issues. Clin Infect Dis 1998; 26:259 - 72, quiz 273-4; http://dx.doi.org/10.1086/516315; PMID: 9502438
- Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 2011; 9:109 - 18; http://dx.doi.org/10.1038/nrmicro2475; PMID: 21189476
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 2005; 15:1150 - 5; http://dx.doi.org/10.1016/j.cub.2005.05.047; PMID: 15964282
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 2004; 3:1076 - 87; http://dx.doi.org/10.1128/EC.3.5.1076-1087.2004; PMID: 15470236
- Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, et al. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol 2005; 56:397 - 415; http://dx.doi.org/10.1111/j.1365-2958.2005.04557.x; PMID: 15813733
- Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci U S A 2003; 100:11007 - 12; http://dx.doi.org/10.1073/pnas.1834481100; PMID: 12958213
- Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, Hube B. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol 2007; 63:1606 - 28; http://dx.doi.org/10.1111/j.1365-2958.2007.05614.x; PMID: 17367383
- Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol 2007; 9:2938 - 54; http://dx.doi.org/10.1111/j.1462-5822.2007.01009.x; PMID: 17645752
- Park H, Liu Y, Solis N, Spotkov J, Hamaker J, Blankenship JR, et al. Transcriptional responses of candida albicans to epithelial and endothelial cells. Eukaryot Cell 2009; 8:1498 - 510; http://dx.doi.org/10.1128/EC.00165-09; PMID: 19700637
- Brennan M, Thomas DY, Whiteway M, Kavanagh K. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol Med Microbiol 2002; 34:153 - 7; http://dx.doi.org/10.1111/j.1574-695X.2002.tb00617.x; PMID: 12381467
- Chamilos G, Lionakis MS, Lewis RE, Lopez-Ribot JL, Saville SP, Albert ND, et al. Drosophila melanogaster as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J Infect Dis 2006; 193:1014 - 22; http://dx.doi.org/10.1086/500950; PMID: 16518764
- Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 2009; 8:1750 - 8; http://dx.doi.org/10.1128/EC.00163-09; PMID: 19666778
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 2002; 297:623 - 6; http://dx.doi.org/10.1126/science.1073759; PMID: 12142542
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol 2002; 12:1209 - 14; http://dx.doi.org/10.1016/S0960-9822(02)00928-4; PMID: 12176330
- Jain C, Yun M, Politz SM, Rao RP. A pathogenesis assay using Saccharomyces cerevisiae and Caenorhabditis elegans reveals novel roles for yeast AP-1, Yap1, and host dual oxidase BLI-3 in fungal pathogenesis. Eukaryot Cell 2009; 8:1218 - 27; http://dx.doi.org/10.1128/EC.00367-08; PMID: 19502579
- Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog 2011; 7:e1002074; http://dx.doi.org/10.1371/journal.ppat.1002074; PMID: 21731485
- Darby C, Cosma CL, Thomas JH, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 1999; 96:15202 - 7; http://dx.doi.org/10.1073/pnas.96.26.15202; PMID: 10611362
- Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 1999; 96:47 - 56; http://dx.doi.org/10.1016/S0092-8674(00)80958-7; PMID: 9989496
- Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 1999; 96:715 - 20; http://dx.doi.org/10.1073/pnas.96.2.715; PMID: 9892699
- Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 1999; 96:2408 - 13; http://dx.doi.org/10.1073/pnas.96.5.2408; PMID: 10051655
- Kurz CL, Chauvet S, Andrès E, Aurouze M, Vallet I, Michel GP, et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J 2003; 22:1451 - 60; http://dx.doi.org/10.1093/emboj/cdg159; PMID: 12660152
- Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol 2000; 10:1539 - 42; http://dx.doi.org/10.1016/S0960-9822(00)00830-7; PMID: 11114525
- Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol 2000; 10:1543 - 5; http://dx.doi.org/10.1016/S0960-9822(00)00833-2; PMID: 11114526
- Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci U S A 2002; 99:15675 - 80; http://dx.doi.org/10.1073/pnas.232568599; PMID: 12438649
- Mylonakis E, Idnurm A, Moreno R, El Khoury J, Rottman JB, Ausubel FM, et al. Cryptococcus neoformans Kin1 protein kinase homologue, identified through a Caenorhabditis elegans screen, promotes virulence in mammals. Mol Microbiol 2004; 54:407 - 19; http://dx.doi.org/10.1111/j.1365-2958.2004.04310.x; PMID: 15469513
- Nakagawa Y, Kanbe T, Mizuguchi I. Disruption of the human pathogenic yeast Candida albicans catalase gene decreases survival in mouse-model infection and elevates susceptibility to higher temperature and to detergents. Microbiol Immunol 2003; 47:395 - 403; PMID: 12906099
- Chávez V, Mohri-Shiomi A, Garsin DA. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect Immun 2009; 77:4983 - 9; http://dx.doi.org/10.1128/IAI.00627-09; PMID: 19687201
- Alarco AM, Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol 1999; 181:700 - 8; PMID: 9922230
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 2000; 16:325 - 7; http://dx.doi.org/10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#; PMID: 10669870
- Gauss KA, Nelson-Overton LK, Siemsen DW, Gao Y, DeLeo FR, Quinn MT. Role of NF-kappaB in transcriptional regulation of the phagocyte NADPH oxidase by tumor necrosis factor-alpha. J Leukoc Biol 2007; 82:729 - 41; http://dx.doi.org/10.1189/jlb.1206735; PMID: 17537988
- Zu YL, Qi J, Gilchrist A, Fernandez GA, Vazquez-Abad D, Kreutzer DL, et al. p38 mitogen-activated protein kinase activation is required for human neutrophil function triggered by TNF-alpha or FMLP stimulation. J Immunol 1998; 160:1982 - 9; PMID: 9469462
- Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell 1997; 90:939 - 49; http://dx.doi.org/10.1016/S0092-8674(00)80358-X; PMID: 9298905
- Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog 2008; 4:e35; http://dx.doi.org/10.1371/journal.ppat.0040035; PMID: 18282097
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, et al. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell 2006; 17:1018 - 32; http://dx.doi.org/10.1091/mbc.E05-06-0501; PMID: 16339080
- Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, et al. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med 2006; 40:1201 - 9; http://dx.doi.org/10.1016/j.freeradbiomed.2005.11.019; PMID: 16545688
- Zhang X, De Micheli M, Coleman ST, Sanglard D, Moye-Rowley WS. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol Microbiol 2000; 36:618 - 29; http://dx.doi.org/10.1046/j.1365-2958.2000.01877.x; PMID: 10844651
- Edens WA, Sharling L, Cheng G, Shapira R, Kinkade JM, Lee T, et al. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J Cell Biol 2001; 154:879 - 91; http://dx.doi.org/10.1083/jcb.200103132; PMID: 11514595
- Hodgkin J, Kuwabara PE, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol 2000; 10:1615 - 8; http://dx.doi.org/10.1016/S0960-9822(00)00867-8; PMID: 11137017
- Vázquez-Torres A, Balish E. Macrophages in resistance to candidiasis. Microbiol Mol Biol Rev 1997; 61:170 - 92; PMID: 9184009
- Arana DM, Alonso-Monge R, Du C, Calderone R, Pla J. Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell Microbiol 2007; 9:1647 - 59; http://dx.doi.org/10.1111/j.1462-5822.2007.00898.x; PMID: 17346314
- Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol 2009; 71:240 - 52; http://dx.doi.org/10.1111/j.1365-2958.2008.06528.x; PMID: 19019164
- Wellington M, Dolan K, Krysan DJ. Live Candida albicans suppresses production of reactive oxygen species in phagocytes. Infect Immun 2009; 77:405 - 13; http://dx.doi.org/10.1128/IAI.00860-08; PMID: 18981256
- Wysong DR, Christin L, Sugar AM, Robbins PW, Diamond RD. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun 1998; 66:1953 - 61; PMID: 9573075
- Reuss O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004; 341:119 - 27; http://dx.doi.org/10.1016/j.gene.2004.06.021; PMID: 15474295
- Bolm M, Jansen WT, Schnabel R, Chhatwal GS. Hydrogen peroxide-mediated killing of Caenorhabditis elegans: a common feature of different streptococcal species. Infect Immun 2004; 72:1192 - 4; http://dx.doi.org/10.1128/IAI.72.2.1192-1194.2004; PMID: 14742574
- Pollock JD, Williams DA, Gifford MA, Li LL, Du X, Fisherman J, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet 1995; 9:202 - 9; http://dx.doi.org/10.1038/ng0295-202; PMID: 7719350
- Jackson SH, Gallin JI, Holland SM. The p47phox mouse knock-out model of chronic granulomatous disease. J Exp Med 1995; 182:751 - 8; http://dx.doi.org/10.1084/jem.182.3.751; PMID: 7650482
- Martel C, Mollin M, Beaumel S, Brion JP, Coutton C, Satre V, et al. Clinical, Functional and Genetic Analysis of Twenty-Four Patients with Chronic Granulomatous Disease - Identification of Eight Novel Mutations in CYBB and NCF2 Genes. J Clin Immunol 2012; 32:942 - 58; http://dx.doi.org/10.1007/s10875-012-9698-8; PMID: 22562447
- Winkelstein JA, Marino MC, Johnston RB Jr., Boyle J, Curnutte J, Gallin JI, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 2000; 79:155 - 69; http://dx.doi.org/10.1097/00005792-200005000-00003; PMID: 10844935
- Aratani Y, Kura F, Watanabe H, Akagawa H, Takano Y, Suzuki K, et al. Critical role of myeloperoxidase and nicotinamide adenine dinucleotide phosphate-oxidase in high-burden systemic infection of mice with Candida albicans. J Infect Dis 2002; 185:1833 - 7; http://dx.doi.org/10.1086/340635; PMID: 12085336
- Brothers KM, Newman ZR, Wheeler RT. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot Cell 2011; 10:932 - 44; http://dx.doi.org/10.1128/EC.05005-11; PMID: 21551247
- Alarco AM, Balan I, Talibi D, Mainville N, Raymond M. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J Biol Chem 1997; 272:19304 - 13; http://dx.doi.org/10.1074/jbc.272.31.19304; PMID: 9235926
- Martchenko M, Alarco AM, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol Biol Cell 2004; 15:456 - 67; http://dx.doi.org/10.1091/mbc.E03-03-0179; PMID: 14617819
- Sherman F, Fink GR, Hicks JB. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, NY, 1986.
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77:71 - 94; PMID: 4366476
- Lay J, Henry LK, Clifford J, Koltin Y, Bulawa CE, Becker JM. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect Immun 1998; 66:5301 - 6; PMID: 9784536
- Sundstrom P, Cutler JE, Staab JF. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect Immun 2002; 70:3281 - 3; http://dx.doi.org/10.1128/IAI.70.6.3281-3283.2002; PMID: 12011025
- Brand A, MacCallum DM, Brown AJ, Gow NA, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell 2004; 3:900 - 9; http://dx.doi.org/10.1128/EC.3.4.900-909.2004; PMID: 15302823
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 1987; 154:164 - 75; http://dx.doi.org/10.1016/0076-6879(87)54076-9; PMID: 3323810
- Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci U S A 2010; 107:1594 - 9; http://dx.doi.org/10.1073/pnas.0912427107; PMID: 20080646
- Ramírez MA, Lorenz MC. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot Cell 2007; 6:280 - 90; http://dx.doi.org/10.1128/EC.00372-06; PMID: 17158734
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998; 14:115 - 32; http://dx.doi.org/10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2; PMID: 9483801
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics 1993; 134:717 - 28; PMID: 8349105
- White TC. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 1997; 41:1482 - 7; PMID: 9210670
- Moran GP, Sullivan DJ, Henman MC, McCreary CE, Harrington BJ, Shanley DB, et al. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother 1997; 41:617 - 23; PMID: 9056003
- Kuhn DM, Mikherjee PK, Clark TA, Pujol C, Chandra J, Hajjeh RA, et al. Candida parapsilosis characterization in an outbreak setting. Emerg Infect Dis 2004; 10:1074 - 81; http://dx.doi.org/10.3201/eid1006.030873; PMID: 15207060
- Laffey SF, Butler G. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology 2005; 151:1073 - 81; http://dx.doi.org/10.1099/mic.0.27739-0; PMID: 15817776
- Mundy RD, Cormack B. Expression of Candida glabrata adhesins after exposure to chemical preservatives. J Infect Dis 2009; 199:1891 - 8; http://dx.doi.org/10.1086/599120; PMID: 19426114