Abstract
A comparative study about protein secretion, immunogenicity and virulence was performed in order to characterize and to compare eight Sporothrix schenckii sensu stricto isolates. For virulence characterization, a murine model, based on survival assay and CFU counting was used. S. brasiliensis and S. globosa, a highly virulent and a non-virulent isolates, respectively were used as external controls. Exoantigen profiles showed different secreted molecules; the 46- and 60-kDa molecules were commonly secreted by all three species. The S. schenckii s. str. isolates could be classified as non-virulent or presenting low, medium or high virulence, based on survival times after infection and recovery of viable fungi. The humoral response profiles of mice infected with S. schenckii s. str., S. globosa and S. brasiliensis were heterogeneous; five virulent isolates (S. schenckii s. str., n = 4 and S. brasiliensis, n = 1) had in common the recognition of the 60-kDa molecule by their respective antisera, suggesting that this antigen may be involved in virulence. Furthermore, the 110-kDa molecule was secreted and recognized by antisera from four virulent isolates (S. schenckii s. str., n = 3 and S. brasiliensis, n = 1), so there is a possibility that this molecule is also related to virulence. Our findings reveal different degrees of virulence in S. schenckii s. str. isolates and suggest the correlation of protein secretion and immunogenicity with virulence of S. schenckii complex. These findings provide new insights into the pathogenesis of S. schenckii s. str. and improve the knowledge about immunogenicity and protein profiles in S. schenckii complex.
Introduction
The genus Sporothrix consists of a rich-species taxon of Ascomycete, which vary according to the ecological niche, frequency, distribution and virulence.Citation1-Citation4 Some of these fungi present potential to survival in mammalian hosts being able to cause damage in multiple species of animals including humans. Since 1898 when the etiological agent of sporotrichosis, Sporothrix schenckii sensu lato (s.l.), was described,Citation5,Citation6 many studies have demonstrated high genetic diversity among different clinical isolates.Citation7-Citation9 Marimon et al. (2006)Citation10 demonstrated that the fungus was actually a complex of cryptic species,Citation10 afterward separated into different phylogenetic speciesCitation11,Citation12 including S. schenckii s. str., S. brasiliensis, S. globosa, S. mexicana and S. luriei. In addition, S. pallida (formerly S. albicans) is a non-pathogenic species closely related to S. schenckii.Citation13 Sporotrichosis is the most common subcutaneous fungal infection found in South America and is usually acquired by traumatic inoculation of saprophytic fungal propagules into subcutaneous tissue, where this thermodimorphic fungus changes its morphology to the parasitic yeast form.Citation14,Citation15 In Brazil, the incidence of sporotrichosis is on rise over the last two decades and it has been attributed to zoonotic outbreaks in cats.Citation16-Citation19 Patients may present several different clinical forms ranging from fixed cutaneous and lymphocutaneous to disseminated form.Citation20 Disseminated sporotrichosis is the most severe infection that has been reported with increasing frequency in recent years mainly in immunosuppressed patients.Citation21-Citation26
Many studies have been conducted to better understand the pathogenesis of sporotrichosis in experimental infection. The decrease in cellular immune responseCitation27 and IL-1 and TNF-α levelsCitation28 between the fourth and sixth week post-infection were reported, thereby providing the colonization of some organs of infected mice by the fungus.Citation29 The IL-4 interleukin is predominantly produced in the fifth week, suggesting the prevalence of humoral immune response in the advanced infection.Citation30
On the other hand, other studies showed the differences in Sporothrix virulence and the clinical presentation of the disease, geographic origin, genotype and thermotolerance.Citation31-Citation34 The time of cultivation of yeast and mycelial cells of S. schenckii induces changes in cell wall composition, causing a decrease of pathogenic power.Citation35 Some comparative studies were conducted correlating the route of experimental infectionCitation32 or clinical manifestation in humans with the virulence degree of different isolates.Citation33,Citation34 Recently, Arrillaga-Moncrieff et al.Citation2,Citation3 showed different degrees of virulence among S. schenckii s. str., S. brasiliensis and S. globosa species.
The aim of this study was to characterize the virulence of eight S. schenckii s. str. isolates compared with S. brasiliensis and S. globosa isolates by the combination of data from CFU assays, survival times and immunoblot assays. Also, the protein secretion of these different isolates will be correlated with humoral response of infected mice in order to find possible antigenic molecules that could be proposed as a possible marker of virulence.
Results
Phenotypic and genotypic characterization of Sporothrix spp isolates
The macroscopic and microscopic morphologies of the S. brasiliensis, S. globosa and S. schenckii s. str. isolates identified herein were very similar to the corresponding reference type strains CBS 120339, CBS 120340 and CBS 359.36, respectively. The only discrepancy that we found was for the isolate Ss51, which presented similarity to the physiological profile of S. brasiliensis, once it did not assimilate sucrose; nevertheless it was identified as S. schenckii s. str. by phylogenetic analysis (described below). Due to this divergence, the phylogenetic analysis was considered to be more consistent for identification of Sporothrix species.
The amplified CAL genes and ITS region yielded DNA fragments of approximately 800bp and 620bp in size, respectively. BLAST searches using CAL and ITS region sequences as a query revealed that seven Brazilian isolates (Ss16, Ss39, Ss40, Ss47, Ss126, Ss141 and Ss143) had a high level (99–100%) of similarity with S. schenckii reference strains. The exception was Ss51, which had 96% similarity with S. brasiliensis IPEC 17920, AM116888). Ss06 (CBS 132922) had high similarity with the type strain of S. globosa (99%, AM116908) and Ss54 (CBS 132990) with S. brasiliensis (100%, AM116899).
Similarity of CAL gene and ITS region among isolates
The CAL and ITS sequences used as reference for phylogenetic analysis were selected based on sequences deposited by De Beer et al.,Citation36 Kawasaki et al.,Citation37 Watanabe et al.,Citation38 Aghayeva et al.,Citation39 Marimon et al.,Citation11,Citation12 Gujjari et al.,Citation40 De Meyer et al.,Citation41 Madrid et al.,Citation42 Romeo et al.Citation13 and Rodrigues et al.Citation4 Both sequences, ITS and CAL loci of the same reference isolate, were not always available, thus, the data were analyzed separately.
The 55 sequences used in CAL data set () were distributed into six main groups, all of them identified in previous studies.Citation11-Citation13 Clades 1–3 and 6 (S. brasiliensis,Citation11 S. schenckii,Citation11 S. globosaCitation11 and S. luriei,Citation12 respectively) are largely composed of pathogens of mammalians whereas groups 4 and 5 (S. mexicanaCitation11 and S. pallida, respectively) are mainly recovered from environmental samples.
Figure 1. Molecular phylogenetic tree generated in this study by Maximum Likelihood based on Kimura 2- parameter (K2P + G) for CAL data set (A) and Tamura 3-parameter (T92 + I) for ITS1/2 + 5.8S data set (B) of S. schenckii complex species. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (Bootstrap support values > 70% are indicated in bold). GenBank accession numbers are indicated next to strain code. Strains used in this study are indicated in bold type.
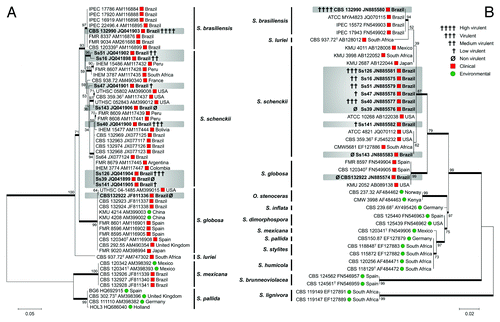
The 39 sequences used in ITS data set () were distributed into 13 main groups, comprising the same four clinical clades described in the CAL data set, as well as the close related environmental Sporothrix species complex and Ophiostoma stenoceras isolates.
The eight isolates (Ss16, Ss39, Ss40, Ss47, Ss51, Ss126, Ss141 and Ss143) clustered significantly into the S. schenckii clade in both analyses. The phylogenetic analysis showed that Ss51 co-clustered in the S. schenckii s. str. clade (reference strain CBS 359.36) in both analyses, confirming its identification as S. schenckii s. str. despite physiological characteristics that were more similar to S. brasiliensis as described above.
The Brazilian isolate Ss06 (CBS 132922) clustered with the S. globosa type strain (CBS 120340) described by Marimon et al.,Citation11 while the feline isolate Ss54 (CBS 132990) clustered with the S. brasiliensis reference strains.
Survival of BALB/c mice following inoculation with Sporothrix spp yeast cells
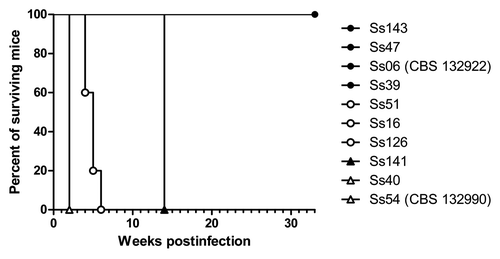
Different times of survival were observed among animals infected with S. schenckii s. str. isolates. The respective survival curves are shown in . All the animals infected with the Ss39, Ss47 and Ss143 isolates survived until the end of the experiment as observed for animals infected with Ss06 (CBS 132922) (S. globosa, control group of non-virulence profile). The isolates Ss16, Ss51 and Ss126 caused the death of all of the mice between fourth and sixth week and Ss141 isolate killed the infected animals at fourteenth week. There were no significant differences (p > 0.05) among Ss16, Ss51, Ss126 and Ss141 but they were statistically different from Ss40 and Ss54 (CBS 132990) (S. brasiliensis, control group of high virulence profile) which killed mice in the second week of infection (p < 0.05). All controls survived until the end of the experiment (data not shown).
Number of viable Sporothrix spp cells recovered from organs
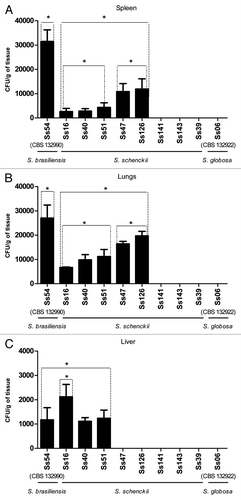
Fungal tissue burden results are shown in . All virulent isolates colonized lungs and spleen. The fungal loads found in spleen and lungs of mice infected with S. brasiliensis (Ss54, CBS 132990) were significantly higher (p < 0.05) than that of S. schenckii s. str. and S. globosa (Ss06, CBS 132922). The fungal loads of spleen and lungs of Ss16, Ss40 and Ss51 were significantly different of Ss47 and Ss126. Interestingly, Ss16 showed remarkable difference of colonization of liver, being significantly higher than Ss54 (CBS 132990) (S. brasiliensis) and other S. schenckii s. str. isolates and S. globosa (p < 0.05). S. globosa were not recovered from any evaluated organs. Taken together the data of survival and CFU (colony forming units) analysis, we classified the virulence degree of each isolate of S. schenckii s. str. based on the profiles of control isolates (Ss54, CBS 132990, and Ss06, CBS 132922). These data showed that S. schenckii s. str. isolates present different degrees of virulence. Thus, Ss40 and Ss126 isolates were considered more virulent among S. schenckii s. str. isolates, followed by Ss16 and Ss51 as medium virulence, Ss47 and Ss141 as low virulence and Ss39 and Ss143 as non-virulent. No fungal load was observed in organs from animals that received sterile PBS (data not shown).
Figure 3. Colony count (CFU/g) comparison among different strains of S. schenckii s. str., and S. brasiliensis or S. globosa in spleen (A), lungs (B) and liver (C) obtained from infected BALB/c mice. Wide bars represent the mean CFU/g of tissue recovered from five mice and thin bars represent the standard deviation. *p < 0.05.
Protein/glycoprotein profile of exoantigens of S. schenckii s. str., S. brasiliensis and S. globosa.
The protein profiles of exoantigens obtained from S. schenckii s. str., S. brasiliensis and S. globosa isolates were heterogeneous ( and ). shows the molecules secreted by each isolate. The 60- and 46-kDa molecules were secreted by all isolates, including S. brasiliensis and S. globosa isolates, and 35-kDa, 80-kDa and 90-kDA molecules were secreted by most of them (). As shown in , there was no association between the profile of the secreted molecules and the fungal species or origin.
Figure 4. (A) Protein/glycoprotein profile of exoantigens of S. schenckii, S. brasiliensis and S. globosa, detected by SDS-PAGE. (B) Immunogenic molecules from exoantigens recognized by antisera from infected animals—western blot assay. S. brasiliensis isolate Ss54 (CBS 132990); S. globosa isolate Ss06 (CBS 132922) and S. schenckii s. str. isolates Ss16, Ss39, Ss40, Ss47, Ss51, Ss 126, Ss141 and Ss143.
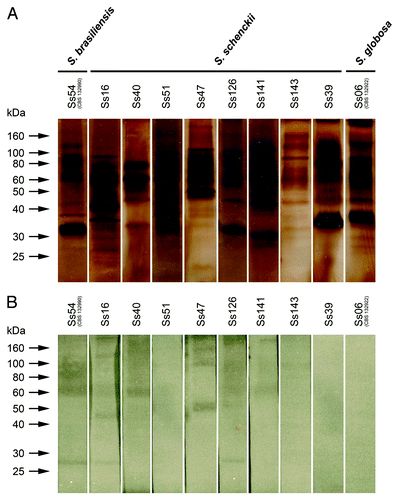
Table 1. Analysis of Sporothrix spp antigens
Western blot: antigen recognition by immune sera
The molecules present in the exoantigens obtained from each isolate and recognized by the respectively antisera from infected mice are shown in and described in . In spite of the 35-kDa and 46-kDa molecules were commonly produced, 46-kDa was immunogenic only for Ss16 and 35-kDa was not recognized by any antisera. Interestingly, all isolates that secreted the 110-kDa molecule, had it recognized by the respective antiserum, including S. brasiliensis isolate. All these isolates presented some pathogenic power. On the other hand, 90-kDa molecule was secreted by six S. schenckii isolates, S. globosa (Ss06) and by S. brasiliensis (Ss54), but only five antisera were able to recognize this molecule, one of them belongs to the non-virulent isolate (Ss143). The 60-kDa molecule was secreted by all isolates and was recognized by four antisera from mice infected with isolates of S. schenckii s. str. that presented some virulence power (Ss16, Ss40, Ss126 and Ss141) and S. brasiliensis isolate (Ss54, CBS 132990). This molecule was not recognized by antisera from non-virulent isolates (Ss143 and Ss39) and S. globosa isolate (Ss06, CBS 132922). Exceptionally, Ss51, a medium virulent isolate, had no molecule recognized by antisera from mice.
Discussion
Recently, several authors proposed that Sporothrix schenckii is indeed a complex of different species that are phylogenetically related and fungi belonging to this genus share typical phenotypic characteristics: S. brasiliensis, S. globosa, S. mexicana, S. schenckii, S. luriei, S. pallida, S. humicola, S. stylites, S. lignivora, S. brunneoviolacea and S. dimorphospora.Citation11,Citation12,Citation41,Citation42 Some studies have investigated the protein secretion profile of Sporothrix,Citation43,Citation44 the genetic variability,Citation11,Citation12,Citation41,Citation42 as well as virulence of some new species.Citation2,Citation3 This is the first study which evaluates the protein secretion of S. brasiliensis, S. globosa and S. schenckii s. str. and connects the humoral immune response to virulence of these species.
The taxonomy was accessed by phenotyping and genotyping assays, using the key features for Sporothrix species differentiation previously proposed by Marimon et al.Citation11,Citation12 and partial sequencing of the CAL gene and ITS region. We, thus, identified one isolate of S. brasiliensis (Ss54, CBS 132990) and one isolate of S. globosa (Ss06, CBS 132922), as well as eight isolates of S. schenckii (Ss16, Ss39, Ss40, Ss47, Ss51, Ss126, Ss141 and Ss143). The characteristics of these species are in agreement with those proposed by Marimon et al.,Citation11 except for Ss51, which was identified as S. schenckii by phylogenetic analysis but did not assimilate sucrose, a key physiological characteristic of S. brasiliensis species. The results of our phylogenetic analysis of the CAL locus are in agreement with those obtained by others.Citation11-Citation13 The phylogenetic relationship using ITS sequence was established among our clinical isolates and some new environmental species recently proposed in the literature belonging to the genus Sporothrix. Sporothrix schenckii s. str. formed a well-supported cluster close related to its sister taxa S. brasiliensis, S. globosa and S. luriei creating a well-supported clinical clade what is in agreement with De Beer et al.Citation36
Various studies using animal models have shown differences in virulence among Sporothrix isolates, correlating the pigmentation of conidia, thermotolerance,Citation31 routes of infectionCitation32 and originCitation33,Citation34 to the degree of virulence, nevertheless, none of these studies take into consideration the new taxonomy used for the clinical S. schenckii complex species.Citation11,Citation12 In this study we did not find correlation between virulence profile of our isolates with thermotolerance or geographic origin (data obtained in our previously studyCitation9) of them. Recently, Arrillaga-Moncrieff et al.Citation2,Citation3 using an immunocompetent murine model, found that S. brasiliensis was the most virulent species, followed by S. schenckii and S. globosa, S. mexicana and S. pallida, which lesional mechanisms could be species specific. They observed that S. brasiliensis presented a remarkable brain tropism while in S. schenckii and S. globosa this tropism was less intense. Part of our results are consistent with these findings because some isolates of S. schenckii s. str. (Ss16, Ss40, Ss47, Ss51, Ss126 and Ss141) are virulent, but others such as Ss39 and Ss143 are non-virulent such as S. globosa, S. mexicana or S. pallida species as demonstrated by Arrillaga-Moncrieff et al.Citation2,Citation3 These non-virulent strains (Ss39 and Ss143) were isolated from patients, and perhaps this fact could be attributed to an underlying disease in the host immunological system.
A great deal of heterogeneity of virulence among the different isolates of S. schenckii s. str. species was observed. To classify the virulence level of each isolate, the results of CFU assay and survival times were combined using as parameters S. brasiliensis (Ss54, CBS 132990) and S. globosa (Ss06, CBS 132922) profiles. The isolate Ss54 (CBS 132990) was the most virulent strain, inducing mortality in a short time of infection, inducing high fungal loads mainly in lungs and spleen and colonizing the three evaluated organs. On the other hand, Ss06 (CBS 132922) was totally non-virulent, because no fungal was recovered from any organ and infected mice survived to the end of the experiment. The virulence profiles of Ss54 (CBS 132990) (S. brasiliensis) and Ss06 (CBS 132922) (S. globosa), here observed, are in agreement with the literature.Citation2,Citation3 For S. schenckii s. str. isolates, all virulent isolates (Ss16, Ss40, Ss51, Ss47 and Ss126) could colonize lungs and spleen and part of them (Ss16, Ss40 and Ss51) were able to disseminate and colonize liver but in low intensity which was similar to S. brasiliensis (Ss54, CBS 132990) colonization. The exception was only Ss16, which fungal loads of liver were significantly higher than Ss54. Based on control group profiles of CFU and survival analysis, Ss40 and Ss126 were classified as high virulence, followed by Ss16 and Ss51 as medium virulence, Ss47 and Ss141 as low virulence and Ss143 and Ss39 as non-virulent isolates. Although Ss40 and Ss126 isolates were considered as high virulent among S. schenckii s. str. isolates, they were less virulent than S. brasiliensis (Ss54, CBS 132990) isolate. In general, the virulence profiles of isolates obtained by CFU and survival assay were in agreement, except for Ss40, Ss47 and Ss141. The Ss40 isolate induced medium fungal loads of all the analyzed organs but the higher inoculum of yeast cells it could induce high mortality as S. brasiliensis (Ss54, CBS 132990). High fungal loads of Ss47 of lungs and spleen were recovered but all infected mice survived to the end of the survival experiment. Taken together, these data show that Ss47 isolate is able to infect mice but these animals, over a long time, can resolve the infection. Ss141 isolate had the opposite behavior, being unable to colonize any analyzed organ, but was able to cause infection when a high inoculum was employed, killing infected animals. These data suggest that a high inoculum of this isolate and a longer infection time is necessary for the consolidation of infection.
In our previous study about virulence factors of Sporothrix spp,Citation9 Ss126 (S. schenckii s. str.) showed proteinase, caseinase, gelatinase, urease and DNase activities. On the other hand, Ss54 (CBS 132990) showed DNase and urease activity.Citation9 Taken together the data of virulence factorsCitation9 and virulence profile (current study), Ss54 expressed less virulence factorsCitation9 than Ss126, but it is more virulent; showing that the mechanisms of pathogenesis are much more complex, involving these virulence factors and others molecules for evasion of immune system.
We found that the 60- and 46-kDa molecules were secreted by all of the isolates, including S. brasiliensis and S. globosa isolates, and thus probably represent important components that are common to all of the studied species. However, the 60-kDa molecule was recognized by five different antisera (one anti-S. brasiliensis and four anti-S. schenckii sera), and the 46-kDa molecule was recognized by only one antisera (anti-S. schenckii). All isolates that had the 60-kDa molecule recognized by antisera from infected mice could kill them. Ss51 (S. schenckii) isolate was able to kill infected animals, but no molecules were recognized by antisera from them. This isolate secreted the 60-kDa molecule, but the molecule was not recognized by the corresponding antiserum; however it was able to kill infected mice. One hypothesis that may explain this behavior could be related to possible post-translational modifications in the 60-kDa molecule among different isolates and thus its antigenicity varies, with some isoforms not eliciting a humoral response in infected mice. On the other hand, sera from mice infected with non-virulent isolates did not recognize the 60-kDa molecule. These data suggest that the 60-kDa molecule may have an influence on the virulence of the isolate that is independent of the species. Further studies are required to investigate the association between this molecule and fungal virulence. The 60-kDa molecule is a possible candidate for being the immunodominant molecule in the S. schenckii complex, although, future studies should be conducted to confirm this hypothesis.
Notwithstanding, the 90-kDa molecule was secreted by eight isolates, only five antisera recognized it. Moreover, the antiserum from Ss143 (non-virulent isolate) recognized this molecule, therefore, this molecule should not be crucial in the pathogenesis. The 110-kDa molecule was secreted by four isolates, and was recognized by their respective antisera. All of these isolates were virulent, so there is a possibility that this molecule is also related to mechanism of virulence.
It is important to know the virulence profile and immunogenicity of the S. schenckii complex for understanding the pathogenesis of sporotrichosis. This is the first study that shows different degrees of virulence among S. schenckii s. str. isolates and a common immunogenic molecule of the S. schenckii complex secreted by most of virulent isolates of S. schenckii and S. brasiliensis. These data bring new insights into the pathogenesis of S. schenckii complex.
Materials and Methods
Characterization of Sporothrix species isolates
The main characteristics of representative Brazilian isolates of S. brasiliensis (Ss54, CBS 132990), S. globosa (Ss06, CBS 132922), and S. schenckii s. str. (Ss16, Ss39, Ss40, Ss47, Ss51, Ss126, Ss141 and Ss143) used in the present study are shown in .
Table 2. Characteristics of Sporothrix spp isolates
We evaluated the fungal growth in potato dextrose agar medium (PDA) at different temperatures (30, 35 and 37°C) by colony diameter measurements and carbohydrate (dextrose, sucrose, raffinose, and ribitol) assimilation according to the phenotyping characterization proposed by Marimon et al.Citation11,Citation12 The characteristics of each isolate were used as taxonomic parameters and applied to a dichotomous key of species in the S. schenckii complex. For phenotyping characterization, we used several S. schenckii complex as reference strains which were previously described by Marimon et al.,Citation11,Citation12 including S. brasiliensis CBS 120339 and IPEC 16919, S. globosa CBS 120340 and FMR 8595 and S. schenckii CBS 359.36 and UTHSC 99-173.
For molecular characterization, the calmodulin (CAL) gene fragment was amplified directly from genomic DNA using the primers CL1 and CL2A.Citation45 In addition, the ITS region was sequenced using primers ITS1 and ITS4, which amplify the 18S, ITS1, 5.8S, ITS2 and 28SCitation46 in order to verify if these isolates were similar to other environmental Sporothrix isolates described by De Meyer et al.Citation41 and Madrid et al.Citation42
Amplified products were gel purified with the Wizard SV Gel and PCR Clean-Up System (Promega Corp.), following the manufacturer’s instructions. DNA samples were completely sequenced with a MegaBACE 1000 DNA Sequencer (Amersham) using the DYEnamic ET Dye Terminator Kit (with Thermo Sequenase II DNA Polymerase). The fragments were sequenced on both strands to increase the quality of sequence data (phred > 30). Sequence alignment was performed using the ClustalW algorithmCitation47 implemented with BioEdit software;Citation48 retrieved alignments were manually corrected to avoid mispaired bases. Nucleotide sequences were exported as FASTA file for BLAST search (www.ncbi.nlm.nih.gov/BLAST).
Phylogenetics analysis
Calmodulin sequences (n = 55) from other isolates of S. schenckii (n = 25), S. brasiliensis (n = 8), S. mexicana (n = 5), S. globosa (n = 12), S. pallida (n = 4) and S. luriei (n = 1), as well as ITS sequences (n = 39) from S. schenckii (n = 15), S. brasiliensis (n = 4), S. mexicana (n = 1), S. globosa (n = 4), S. pallida (n = 1), S. luriei (n = 1), S. inflata (n = 1), S. dimorphospora (n = 2), S. stylites (n = 2), S. humicola (n = 2), S. brunneoviolacea (n = 2), S. lignivora (n = 2) and Ophiostoma stenoceras (n = 2) were included in the present study as reference strains for the phylogenetic analysis. These sequences were previously deposited at GenBank (www.ncbi.nlm.nih.gov/genbank) and described by De Beer et al.,Citation36 Kawasaki et al.,Citation37 Watanabe et al.,Citation38 Aghayeva et al.,Citation39 Marimon et al.,Citation11,Citation12 Gujjari et al.,Citation40 De Meyer et al.,Citation41 Madrid et al.,Citation42 Romeo et al.Citation13 and Rodrigues et al.Citation4 Evolutionary analyses were conducted in MEGA5Citation49 with Maximum Likelihood method. Evolutionary distances were computed using the Kimura 2-parameter methodCitation50 with a discrete Gamma distribution for the CAL data set and Tamura 3-parameter methodCitation51 for ITS data set, with 1,000 bootstrap replicates.Citation52
Exoantigens preparation
Exoantigens from mycelial phase of S. schenckii s. str., S. globosa and S. brasiliensis isolates were obtained as previous describedCitation43 using Sabouraud medium (Difco). Protein content was determined according to the Bradford method.Citation53
Experimental infection in mice
Preparation of Sporothrix spp yeast cells for inoculum
Our methods were based on those described by Carlos et al.Citation29 Each isolate was initially grown in brain-heart-infusion agar slants (BHI) (Difco) until complete reversion to the yeast phase. Then, the total growth of three slants was transferred to 300 ml of BHI broth and incubated at 37°C for 7 d in a rotatory shaker with constant agitation (100 rpm/min). Yeast cultures were washed three times with phosphate-buffered saline (PBS), centrifuged at 700 × g for 15 min, and finally suspended in sterile PBS. The yeast cells concentration was adjusted to 1 × 106 or 5 × 106 after counting with a hemocytometer. The viability of these inocula was verified by plating dilutions of the suspension on BHI plates.
Virulence assays
Animals
A total of 165 male, 6–8 weeks old BALB/c mice, were purchased from Federal University of São Paulo (UNIFESP) for virulence assays of which 55 mice were used for CFU assay and 110 mice were used for survival assay. Mice were housed in temperature-controlled rooms at 23–25°C with ad libitum access to food and water. All the procedures were approved by UNIFESP Ethics Committee and are in accordance with the National Institutes of Health Animal and Care Guidelines.
Mice survival assay
Male BALB/c mice (n = 110) were divided into 11 groups of ten mice each (one group for each fungal strain and one negative control group). The mice were anesthetized with 0.2 mg/kg xilazine and 20 mg/kg ketamine and inoculated i.v. with 5 × 106 fungal yeast cells/animal. Control groups were injected with 100 µl of PBS. Mice were observed daily over 31 weeks, and deaths were recorded daily.
CFU analysis and sera obtaintion
Male BALB/c mice (n = 55) were divided into 11 groups of 5 mice per group (one group for each fungal strain and one negative control group). The mice were anesthetized with 0.2 mg/kg xilazine and 20 mg/kg ketamine and inoculated intravenously (i.v.) with 1 × 106 yeast cells/animal.Citation29 Animals from negative control group received 100 µl of sterile PBS. On the fifth week after infection,Citation29 the animals were sacrificed by CO2 anesthesia, and the spleen, lungs and liver were aseptically removed. The organs were separated, weighed and homogenized in sterile PBS using a tissue grinder. Samples (100 μl) of each homogenate were seeded on Petri dishes containing BHI agar and incubated at 37°C. Colonies were counted from the seventh day until the fifteenth day. The results were expressed as colony-forming units (CFU)/g tissue. Sera from mice were collected and kept at −20°C for western blot assay.
Statistical analysis
For CFU assays, comparisons between groups were analyzed by analysis of variance (ANOVA) followed by post-hoc Tukey, with significance assumed to be p < 0.05. For survival assays, data were analyzed by Kaplan-Meier survival plots followed by log-rank test. Data with a p value < 0.05 or less were considered to be significant.Citation54
SDS-PAGE and western blot assay
Exoantigens (2 µg of protein) were analyzed by SDS-PAGE using 10% gels, as described by Laemmli et al.Citation55 The SDS-PAGE was stained with silver stain, and the relative molecular weights of the fractions were estimated using standard broad range molecular weight markers (Benchmark Invitrogen). For western blot assay, 5 µg per slot of protein were applied into SDS-PAGE and electrotransferred onto nitrocellulose membranes (Bio-Rad Laboratories, Inc.) (0.45-µm pore size) in a transblotting chamber (Biorad MiniPROTEAN Tetra Cell, Bio-Rad Laboratories, Inc.) using Tris-glycine-methanol buffer (pH 8.3)Citation56 for 16 h at 14 mA at 4°C. The membranes were incubated in PBS containing 1% bovine serum albumin for 4 h at 37°C. The membrane was then cut lengthwise into 0.5-cm strips, and each strip was placed in a separate tray and incubated with 1:50 dilution (in PBS-Tween 20, 0.005%) of different mice sera for 1 h at room temperature (RT). The assay was done with sera from all of the mice from all of the experimental groups (serum vs. exoantigen from the same isolate). The membrane strips were washed three times in PBS-Tween 20, 0.005%) and then incubated for 1 h at RT with a 1:1,000 dilution of peroxidase-conjugated IgG goat anti-mouse IgG (Sigma-Aldrich Co.). After additional washes, the membrane strips were incubated with a substrate that consisted of 5 mg of DAB (3,3′-diaminobenzidine), 50 ml of 1 × PBS and 90 µl of hydrogen peroxide. After color development, the membrane strips were washed with distilled water and dried. presents a combined analysis of SDS-PAGE and western blot results.
Acknowledgments
This study was supported by FAPESP (2009/54024-2). G.F.F. (2011/01628-8) and A.M.R. (2011/07350-1) are fellows of FAPESP.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- De Hoog GS. The genera Blastobotrys, Sporothrix, Calcarisporium and Calcarisporiella gen. nov.. Stud Mycol 1974; 7:1 - 84
- Arrillaga-Moncrieff I, Capilla J, Mayayo E, Marimon R, Mariné M, Gené J, et al. Different virulence levels of the species of Sporothrix in a murine model. Clin Microbiol Infect 2009; 15:651 - 5; http://dx.doi.org/10.1111/j.1469-0691.2009.02824.x; PMID: 19624508
- Arrillaga-Moncrieff I, Capilla J, Fernández AM, Fariñas F, Mayayo E. Diferencias en la patogenicidad del complejo de especies Sporothrix en un modelo animal. Patologia 2010; 48:82 - 7
- Rodrigues AM, de Hoog S, de Camargo ZP. Emergence of pathogenicity in the Sporothrix schenckii complex. Med Mycol 2012; In press http://dx.doi.org/10.3109/13693786.2012.719648; PMID: 22989196
- Schenck BR. On refractory subcutaneous abscesses caused by a fungus possibly related to sporothrichia. Bull Johns Hopkins Hosp 1898; 9:286 - 90
- Hektoen L, Perkins CF. Refractory subcutaneous abscesses caused by Sporothrix schenckii. A new pathogenic fungus. J Exp Med 1900; 5:77 - 89; http://dx.doi.org/10.1084/jem.5.1.77; PMID: 19866937
- Mesa-Arango AC, Del Rocío Reyes-Montes M, Pérez-Mejía A, Navarro-Barranco H, Souza V, Zúñiga G, et al. Phenotyping and genotyping of Sporothrix schenckii isolates according to geographic origin and clinical form of Sporotrichosis. J Clin Microbiol 2002; 40:3004 - 11; http://dx.doi.org/10.1128/JCM.40.8.3004-3011.2002; PMID: 12149366
- Neyra E, Fonteyne PA, Swinne D, Fauche F, Bustamante B, Nolard N. Epidemiology of human sporotrichosis investigated by amplified fragment length polymorphism. J Clin Microbiol 2005; 43:1348 - 52; http://dx.doi.org/10.1128/JCM.43.3.1348-1352.2005; PMID: 15750106
- Fernandes GF, Santos PO, Amaral CC, Sasaki AA, Martinez-Godoy P, Camargo ZP. Characteristics of 151 Brazilian Sporothrix schenckii isolates from 5 different geographic regions of Brazil: a forgotten and re-emergent pathogen. Open Mycol J 2009; 3:48 - 58; http://dx.doi.org/10.2174/1874437000903010048
- Marimon R, Gené J, Cano J, Trilles L, Dos Santos Lazéra M, Guarro J. Molecular phylogeny of Sporothrix schenckii.. J Clin Microbiol 2006; 44:3251 - 6; http://dx.doi.org/10.1128/JCM.00081-06; PMID: 16954256
- Marimon R, Cano J, Gené J, Sutton DA, Kawasaki M, Guarro J. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol 2007; 45:3198 - 206; http://dx.doi.org/10.1128/JCM.00808-07; PMID: 17687013
- Marimon R, Gené J, Cano J, Guarro J. Sporothrix luriei: a rare fungus from clinical origin. Med Mycol 2008; 46:621 - 5; http://dx.doi.org/10.1080/13693780801992837; PMID: 19180753
- Romeo O, Scordino F, Criseo G. New insight into molecular phylogeny and epidemiology of Sporothrix schenckii species complex based on calmodulin-encoding gene analysis of Italian isolates. Mycopathologia 2011; 172:179 - 86; http://dx.doi.org/10.1007/s11046-011-9420-z; PMID: 21461774
- Kwon-Chung K, Bennett J. Chapter 26 - Sporotrichosis. In: Kwon-Chung K, Bennett J, eds. Medical Mycology. 1st ed. Philadelphia: Lea & Febiger, 1992: 707-29.
- Morris-Jones R. Sporotrichosis. Clin Exp Dermatol 2002; 27:427 - 31; http://dx.doi.org/10.1046/j.1365-2230.2002.01087.x; PMID: 12372075
- Schubach TM, Schubach A, Okamoto T, Barros MB, Figueiredo FB, Cuzzi T, et al. Evaluation of an epidemic of sporotrichosis in cats: 347 cases (1998-2001). J Am Vet Med Assoc 2004; 224:1623 - 9; http://dx.doi.org/10.2460/javma.2004.224.1623; PMID: 15154732
- Schubach A, Schubach TM, Barros MB, Wanke B. Cat-transmitted sporotrichosis, Rio de Janeiro, Brazil. Emerg Infect Dis 2005; 11:1952 - 4; http://dx.doi.org/10.3201/eid1112.040891; PMID: 16485488
- Barros MBL, Schubach TP, Coll JO, Gremião ID, Wanke B, Schubach A. [Sporotrichosis: development and challenges of an epidemic]. Rev Panam Salud Publica 2010; 27:455 - 60; PMID: 20721446
- Falqueto A, Bravim Maifrede S, Araujo Ribeiro M. Unusual clinical presentation of sporotrichosis in three members of one family. Int J Dermatol 2012; 51:434 - 8; http://dx.doi.org/10.1111/j.1365-4632.2011.5085.x; PMID: 22435433
- Lopes-Bezerra LM, Schubach A, Costa RO. Sporothrix schenckii and sporotrichosis. An Acad Bras Cienc 2006; 78:293 - 308; http://dx.doi.org/10.1590/S0001-37652006000200009; PMID: 16710567
- Shaw JC, Levinson W, Montanaro A. Sporotrichosis in the acquired immunodeficiency syndrome. J Am Acad Dermatol 1989; 21:1145 - 7; http://dx.doi.org/10.1016/S0190-9622(89)70318-2; PMID: 2808849
- Donabedian H, O’Donnell E, Olszewski C, MacArthur RD, Budd N. Disseminated cutaneous and meningeal sporotrichosis in an AIDS patient. Diagn Microbiol Infect Dis 1994; 18:111 - 5; http://dx.doi.org/10.1016/0732-8893(94)90075-2; PMID: 8062528
- Bunce PE, Yang L, Chun S, Zhang SX, Trinkaus MA, Matukas LM. Disseminated sporotrichosis in a patient with hairy cell leukemia treated with amphotericin B and posaconazole. Med Mycol 2012; 50:197 - 201; http://dx.doi.org/10.3109/13693786.2011.584074; PMID: 21612561
- Freitas DF, de Siqueira Hoagland B, do Valle AC, Fraga BB, de Barros MB, de Oliveira Schubach A, et al. Sporotrichosis in HIV-infected patients: report of 21 cases of endemic sporotrichosis in Rio de Janeiro, Brazil. Med Mycol 2012; 50:170 - 8; http://dx.doi.org/10.3109/13693786.2011.596288; PMID: 21859385
- Silva-Vergara ML, Maneira FR, De Oliveira RM, Santos CT, Etchebehere RM, Adad SJ. Multifocal sporotrichosis with meningeal involvement in a patient with AIDS. Med Mycol 2005; 43:187 - 90; http://dx.doi.org/10.1080/13693780500035904; PMID: 15832562
- Silva-Vergara ML, de Camargo ZP, Silva PF, Abdalla MR, Sgarbieri RN, Rodrigues AM, et al. Disseminated Sporothrix brasiliensis infection with endocardial and ocular involvement in an HIV-infected patient. Am J Trop Med Hyg 2012; 86:477 - 80; http://dx.doi.org/10.4269/ajtmh.2012.11-0441; PMID: 22403321
- Carlos IZ, Sgarbi DB, Angluster J, Alviano CS, Silva CL. Detection of cellular immunity with the soluble antigen of the fungus Sporothrix schenckii in the systemic form of the disease. Mycopathologia 1992; 117:139 - 44; http://dx.doi.org/10.1007/BF00442774; PMID: 1640975
- Carlos IZ, Zini MM, Sgarbi DB, Angluster J, Alviano CS, Silva CL. Disturbances in the production of interleukin-1 and tumor necrosis factor in disseminated murine sporotrichosis. Mycopathologia 1994; 127:189 - 94; http://dx.doi.org/10.1007/BF01102920; PMID: 7808513
- Carlos IZ, Sgarbi DB, Placeres MC. Host organism defense by a peptide-polysaccharide extracted from the fungus Sporothrix schenckii.. Mycopathologia 1998; 144:9 - 14; http://dx.doi.org/10.1023/A:1006964516334; PMID: 10422268
- Maia DC, Sassá MF, Placeres MC, Carlos IZ. Influence of Th1/Th2 cytokines and nitric oxide in murine systemic infection induced by Sporothrix schenckii.. Mycopathologia 2006; 161:11 - 9; http://dx.doi.org/10.1007/s11046-005-0142-y; PMID: 16389479
- Dixon DM, Salkin IF, Duncan RA, Hurd NJ, Haines JH, Kemna ME, et al. Isolation and characterization of Sporothrix schenckii from clinical and environmental sources associated with the largest U.S. epidemic of sporotrichosis. J Clin Microbiol 1991; 29:1106 - 13; PMID: 1864926
- Tachibana T, Matsuyama T, Mitsuyama M. Characteristic infectivity of Sporothrix schenckii to mice depending on routes of infection and inherent fungal pathogenicity. Med Mycol 1998; 36:21 - 7; http://dx.doi.org/10.1080/02681219880000041; PMID: 9776808
- Kong X, Xiao T, Lin J, Wang Y, Chen HD. Relationships among genotypes, virulence and clinical forms of Sporothrix schenckii infection. Clin Microbiol Infect 2006; 12:1077 - 81; http://dx.doi.org/10.1111/j.1469-0691.2006.01519.x; PMID: 17002606
- Brito MM, Conceição-Silva F, Morgado FN, Raibolt PS, Schubach A, Schubach TP, et al. Comparison of virulence of different Sporothrix schenckii clinical isolates using experimental murine model. Med Mycol 2007; 45:721 - 9; http://dx.doi.org/10.1080/13693780701625131; PMID: 17885952
- Fernandes KS, Mathews HL, Lopes Bezerra LM. Differences in virulence of Sporothrix schenckii conidia related to culture conditions and cell-wall components. J Med Microbiol 1999; 48:195 - 203; http://dx.doi.org/10.1099/00222615-48-2-195; PMID: 9989648
- de Beer ZW, Harrington TC, Vismer HF, Wingfield BD, Wingfield MJ. Phylogeny of the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 2003; 95:434 - 41; http://dx.doi.org/10.2307/3761885; PMID: 21156632
- Kawasaki M, Watanabe S, Ishizaki H. . 2003, http://www.ncbi.nlm.nih.gov/nuccore.
- Watanabe S, Kawasaki M, Mochiduki T, Ishizaki H. . 2004, http://www.ncbi.nlm.nih.gov/nuccore.
- Aghayeva DN, Wingfield MJ, Kirisits T, Wingfield BD. Ophiostoma dentifundum sp. nov. from oak in Europe, characterized using molecular phylogenetic data and morphology. Mycol Res 2005; 109:1127 - 36; http://dx.doi.org/10.1017/S0953756205003710; PMID: 16279407
- Gujjari P, Suh SO, Zhou J. . 2008; http://www.ncbi.nlm.nih.gov/nuccore.
- de Meyer EM, de Beer ZW, Summerbell RC, Moharram AM, de Hoog GS, Vismer HF, et al. Taxonomy and phylogeny of new wood- and soil-inhabiting Sporothrix species in the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 2008; 100:647 - 61; http://dx.doi.org/10.3852/07-157R; PMID: 18833758
- Madrid H, Gené J, Cano J, Silvera C, Guarro J. Sporothrix brunneoviolacea and Sporothrix dimorphospora, two new members of the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 2010; 102:1193 - 203; http://dx.doi.org/10.3852/09-320; PMID: 20943519
- Fernandes GF, Do Amaral CC, Sasaki AA, Godoy PM, De Camargo ZP. Heterogeneity of proteins expressed by Brazilian Sporothrix schenckii isolates. Med Mycol 2009; 47:855 - 61; http://dx.doi.org/10.3109/13693780802713216; PMID: 19184772
- Mendoza M, Díaz AM, Hung MB, Zambrano EA, Díaz E, De Albornoz MC. Production of culture filtrates of Sporothrix schenckii in diverse culture media. Med Mycol 2002; 40:447 - 54; PMID: 12462523
- O'Donnell K. Molecular phylogeny of the Nectria haematococca - Fusarium solani species complex. Mycologia 2000; 92:919 - 38; http://dx.doi.org/10.2307/3761588
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR Protocols – a Guide to Methods and Application. San Diego: Academic Press, 1990:315-22.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673 - 80; http://dx.doi.org/10.1093/nar/22.22.4673; PMID: 7984417
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41:95 - 8
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731 - 9; http://dx.doi.org/10.1093/molbev/msr121; PMID: 21546353
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980; 16:111 - 20; http://dx.doi.org/10.1007/BF01731581; PMID: 7463489
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C content biase. Mol Biol Evol 1992; 4:678 - 87
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985; 39:783 - 91; http://dx.doi.org/10.2307/2408678
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248 - 54; http://dx.doi.org/10.1016/0003-2697(76)90527-3; PMID: 942051
- Zar JH. Biostatistical analysis, 2nded. New Jersey: Prentice- Hall, Englewood Cliffs 1984.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680 - 5; http://dx.doi.org/10.1038/227680a0; PMID: 5432063
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 1979; 76:4350 - 4; http://dx.doi.org/10.1073/pnas.76.9.4350; PMID: 388439