Abstract
The Burkholderia cepacia complex (Bcc) consists of 17 closely related species that are problematic opportunistic bacterial pathogens for cystic fibrosis patients and immunocompromised individuals. These bacteria are capable of utilizing two different chemical languages: N-acyl homoserine lactones (AHLs) and cis-2-unsaturated fatty acids. Here we summarize the current knowledge of the underlying molecular architectures of these communication systems, showing how they are interlinked and discussing how they regulate overlapping as well as specific sets of genes. A particular focus is laid on the role of these signaling systems in the formation of biofilms, which are believed to be highly important for chronic infections. We review genes that have been implicated in the sessile lifestyle of this group of bacteria. The new emerging role of the intracellular second messenger cyclic dimeric guanosine monophosphate (c-di-GMP) as a downstream regulator of the fatty acid signaling cascade and as a key factor in biofilm formation is also discussed.
Introduction
The Burkholderia cepacia complex (Bcc) is a group of closely related species with large genomes comprising multiple replicons. Bcc species are notable for their ability to metabolize a wide range of organic compounds and to thrive in many different environments.Citation1-Citation4 The Bcc currently contains 17 defined species, which include opportunistic pathogens that are best known for infecting immunocompromised patients or individuals suffering from cystic fibrosis (CF), where they can cause a fatal pneumonia known as “cepacia syndrome”.Citation5 One of the major problems associated with Bcc infection is their intrinsic resistance to antibiotic therapy. Several strains of the Bcc species B. multivorans, B. cenocepacia, B. cepacia, and B. dolosa have been shown to be highly transmissible between patients, with B. cenocepacia and B. multivorans accounting for the majority of CF infections.Citation6-Citation8 Biofilm formation is a common trait of Bcc strains and has been associated with the persistence of Bcc infections and the increased resistance to antibiotics relative to planktonic cells.Citation9-Citation13 It has been shown that in many Bcc strains the formation of biofilms, as well as the expression of virulence factors and secondary metabolites, is under quorum sensing (QS) control.Citation14-Citation17 QS is a cell density-dependent regulatory mechanism used by bacteria to coordinate gene expression by the aid of diffusible self-produced signal molecules. The purpose of this review is to summarize the current knowledge of QS in members of the Bcc and discuss its role in biofilm formation and pathogenicity.
AHL-Based QS Systems in the Bcc
All Bcc members encode at least one QS system that consists of homologs of the LuxR and LuxI proteins of Vibrio fischeri, where LuxI synthesizes an AHL signal and LuxR is an AHL receptor protein that activates or represses gene expression by binding to a consensus sequence (the so-called lux box) in the promoter regions of target genes.Citation18 AHL signal molecules can differ in the length and substitution of their acyl side chains. In many cases transcription of luxI is activated by the LuxR/AHL complex, providing a signal amplification mechanism via positive feed-back regulation.Citation19
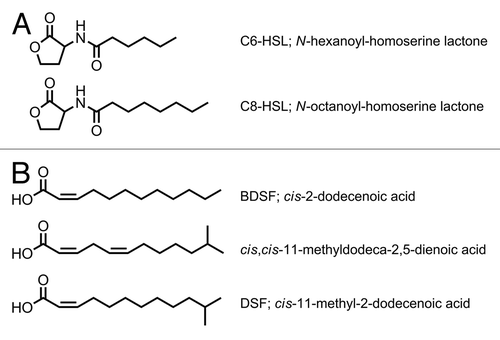
AHL production in the Bcc is strain-dependent with respect to both the quantity and the type of AHL molecules. Within the Bcc the CepIR QS system is fully conserved.Citation20-Citation23 CepI directs the synthesis of N-octanoyl-homoserine lactone (C8-HSL) and minor amounts of N-hexanoyl-homoserine lactone (C6-HSL; ).Citation14,Citation22 Epidemic strains of B. cenocepacia belonging to the ET12 lineage carry the B. cenocepacia genomic island (cci), which encodes an additional QS system named CciIR.Citation24 The AHL synthase CciI produces C6-HSL and minor amounts of C8-HSL, which are bound by the cognate receptor CciR. The CepIR and CciIR systems interact, with CepR positively regulating the expression of the cciIR operon and CciR negatively regulating cepI expression.Citation15 While CepR is mainly a positive regulator, CciR acts as a negative regulator of gene expression. In B. vietnamiensis strains, CepR is required for expression of yet another QS system (BviIR), which utilizes N-decanoyl-homoserine lactone (C10-HSL).Citation25 Phenotypic assays as well as global transcript and protein analyses using cepIR and cciIR mutant strains have shown that AHL-mediated QS controls various functions, including swarming motility, biofilm formation and the production of virulence factors, such as proteases (e.g., the metalloproteases ZmpA and ZmpB), siderophores, toxins, and antifungal agents.Citation14-Citation16,Citation26,Citation27
Figure 1. The different QS molecules produced by Bcc species. (A) Signaling molecules in AHL-based QS. (B) Signaling molecules in DSF family-based QS.
AHL-mediated QS is fine-tuned by additional regulators
The AHL synthase-encoding genes and their cognate receptor genes are normally located in close proximity to each other in the genome. However, whole genome sequencing identified LuxR homologs in significant excess relative to the number of AHL synthases. Although these unpaired, “orphan” or “solo” LuxR proteins are not associated with an AHL synthase, most of them require AHLs in order to function.Citation28-Citation30 An orphan LuxR homolog, CepR2, has been identified in all B. cenocepacia strains sequenced so far but not in other Bcc species. While CepR2 is not involved in the regulation of cepIR or cciIR, CepR is required for CepR2-mediated activation of pyochelin production in B. cenocepacia strain H111.Citation31 In contrast, the CepR2 in B. cenocepacia strain K56-2 acts as a repressor; moreover, in this strain expression of CepR2 is repressed by CciR and its activity is antagonized by AHLs.Citation32 Recently, CepR2 of strain K56-2 has been shown to function as an anti-activator of CepS, an AraC-type regulator-encoding gene located adjacent to cepR2. CepS was shown to act downstream of CepR2 by activating gene expression in the absence of CepR2.Citation32
Additional regulators involved in fine-tuning AHL-mediated QS have been identified in B. cenocepacia. A hypothetical conserved protein encoded by BCAM1871, which is downstream and co-transcribed with cepI, was shown to induce AHL activity and to positively regulate cepIR and cciIR expression.Citation33 In addition, BCAM1871 activates expression of the LysR-type regulator ShvR, which is highly conserved among members of the Bcc and is located in the vicinity of the antifungal cluster (afc) on plasmid pC3 (formerly chromosome 3).Citation33,Citation34 ShvR has been shown to negatively control cepIR and cciIR expression while positively affecting biofilm formation.Citation35,Citation36 Another negative regulator of cepI and cciI expression and their target genes is the recently identified membrane hybrid sensor kinase AtsR (Adhesion and type six secretion system regulator) which is lacking a DNA binding domain, suggesting control through a signal transduction cascade.Citation37 AtsR has been shown to be a global regulator, which controls expression of virulence factors also in the absence of the CepIR QS system. The exact mode of action of AtsR as well as of the other regulators affecting the QS circuitry in Bcc strains remain to be elucidated.
Fatty Acid Signal-Based QS in the Bcc
In 2008, Boon et al. reported the identification of a novel fatty acid signal molecule that is produced by several B. cenocepacia strains.Citation38 The structure of the molecule synthesized by B. cenocepacia J2315 was identified as cis-2-dodecenoic acid, referred to as BDSF (Burkholderia diffusible signal factor). BDSF is structurally related to DSF (diffusible signal factor, cis-11-methyl-2-dodecenoic acid), which was first isolated from supernatants of Xanthomonas campestris pv campestris.Citation39,Citation40 In this plant pathogen DSF is activating the RpfCG two component system, leading to a lowered cellular c-di-GMP level and concomitantly to the expression of target genes involved in biofilm dispersal and virulence.Citation41,Citation42 DSF is synthesized by the gene products of rpfF and rpfB encoding a putative enoyl-CoA hydratase and a putative long-chain fatty acyl CoA ligase, respectively.Citation42,Citation43 In B. cenocepacia BDSF is synthesized by an RpfF homolog (BCAM0581 in B. cenocepacia J2315), named RpfFBc.Citation38,Citation44 RpfFBc is a bifunctional crotonase having both dehydratase and thioesterase activities, which enables the enzyme to directly convert the acyl carrier protein (ACP) thioester of 3-hydroxydodecanoic acid into cis-2-dodecenoic acid.Citation44 BDSF accumulates in a cell density-dependent manner with maximum levels observed in the late stationary phase.Citation45 However, in contrast to the AHL-dependent CepIR system, the expression of the BDSF synthase is not subject to positive feedback regulation.Citation14,Citation45 The biological activity of BDSF is dependent on the cis configuration of the fatty acid, as neither the trans isomer of BDSF nor lauric acid were found to be active.Citation46 Moreover, supplementation of the medium with DSF rescued the phenotypes of an rpfFBc mutant, providing further evidence that the configuration of the double bond is a critical structural feature of the signal molecule. This result also suggests that cis-2-unsaturated fatty acids may be used for interspecies communication. Not only can DSF substitute for BDSF in B. cenocepacia but it has also been shown that BDSF is able to activate DSF-dependent responses in X. campestris, suggesting that cross-species signaling between these organisms is possible.Citation38 In this context it is worth noting that the production of DSF-family signal molecules is widespread among bacteria and therefore cross-talk via cis-2-unsaturated fatty acids may be a common phenomenon.Citation47,Citation48 A recent study showed that DSF-family molecules, including BDSF, are present in sputum taken from CF patients and it was suggested that interspecies DSF-mediated bacterial interactions occur in the CF lung and may influence the efficacy of antibiotic treatment.Citation49 Finally, BDSF has been shown to inhibit germ tube formation of Candida albicans, indicating that BDSF also serves as an interkingdom signal.Citation38,Citation50
The BDSF-regulated QS system is involved in the control of several functions. Mutation of rpfFBc resulted in decreased motility, reduced adherence to porcine mucin, diminished exopolysaccharides (EPS) production and lowered protease activity.Citation46,Citation51,Citation52 In addition, the BDSF mutant strains were found to be more susceptible to antimicrobials and their ability to form biofilms was shown to be strongly reduced.Citation53,Citation54
BDSF perception
Recent work has shown that the gene adjacent to rpfFBc encodes the BDSF receptor protein RpfR (BCAM0580 in B. cenocepacia J2315).Citation46 RpfR contains a GGDEF, an EAL, and a PAS domain. PAS domains are able to bind a chemically diverse range of small-molecules, including hemes, flavins, di- and tricarboxylic acids, divalent metal cations, amino acids and coumaric acid.Citation55 RpfR and RV1364c, a σF regulatory protein of Mycobacterium tuberculosis, are the only proteins known to bind directly a fatty acid through their PAS domains.Citation46,Citation56 GGDEF and EAL are highly conserved domains involved in c-di-GMP turnover with diguanylate cyclase and phosphodiesterase activities, respectively.Citation57 Binding of BDSF to the PAS domain causes an allosteric conformational change of RpfR and thereby stimulates the c-di-GMP phosphodiesterase activity of the protein.Citation46 Consequently, in the presence of BDSF RpfR lowers the intracellular c-di-GMP level. Noteworthy, RpfR is the first example of a c-di-GMP metabolic enzyme that is directly activated by a QS signal molecule.Citation46
McCarthy et al. identified an additional putative BDSF sensor, BCAM0227, in B. cenocepacia J2315.Citation53 This protein shares 35.6% identity with RpfC of X. campestris. BCAM0227 contains a histidine kinase phosphoacceptor domain (HisKA), a CheY-like receiver domain, and a C-terminal histidine phosphotransfer (HPt) domain. However, at least two lines of evidence indicate that the BCAM0227 signaling system may only play a subordinate role in responding to accumulated BDSF signals. First, disruption of BCAM0227 only affected the expression of a subset of genes belonging to the BDSF regulon. Second, while mutation of BCAM0227 and rpfFBc has a similar effect on biofilm architecture, the two mutants differed drastically in motility and in adherence to porcine mucin. In contrast, a rpfR mutation leads to identical phenotypes as rpfFBc mutations, including reduced motility, reduced biofilm formation, lowered proteolytic activity and attenuated virulence.Citation27,Citation46
The BDSF and AHL stimulons overlap
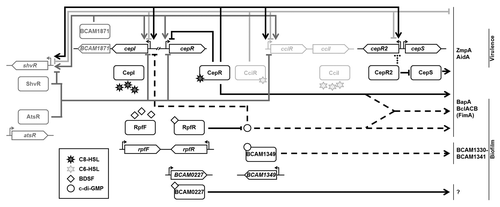
In order to identify BDSF-regulated genes, transposon mutagenesis, global transcript and protein profiling analyses were performed.Citation27,Citation51,Citation53 In B. cenocepacia J2315, rpfFBc was found to regulate 372 genes with functions including motility, attachment, stress tolerance, virulence, transport, signal transduction, multidrug resistance, and detoxification.Citation53 Among these RpfFBc-dependent genes, 65 were found to be also regulated by the putative BDSF sensor BCAM0227. The mapping of the B. cenocepacia H111 BDSF stimulon by RNA-Seq and shotgun proteomics confirmed BDSF-dependent regulation of genes known to be involved in biofilm formation and protease activity.Citation27 Among the genes positively regulated by BDSF, many are known to be controlled by the CepIR QS system, suggesting that both signal molecules are required for full expression of some functions, including the large surface protein BapA and the EPS cepacian.Citation27,Citation58 However, the contribution of each of the systems in the regulation of target genes was found to be variable.Citation27 While some genes are mainly regulated by either BDSF (e.g., bclACB) or AHLs (e.g., aidA or cepI), maximum transcription of bapA requires the presence of both signal molecules. On the basis of these results we proposed a model in which the BDSF and AHL QS systems operate in parallel to regulate specific as well as overlapping sets of genes ().Citation27 BDSF-dependent signaling results in a reduction of the intracellular c-di-GMP levels, which in turn leads to differential expression of target genes.Citation46 Hence, it is very likely that an as-yet unidentified regulator (or regulatory cascade) responds to changes in the intracellular c-di-GMP level. Whether this regulator is independent of the CepIR system or the two QS regulatory cascades converge in one regulator remains to be elucidated.
Figure 2. Model of the QS network in B. cenocepacia. Depicted in black is the “core” QS circuitry of B. cenocepacia that is present in all strains and that consists of the CepIR and the RpfFR systems, as well as the downstream regulators CepR2, CepS, BCAM1349 and BCAM0227. The regulators ShvR, AtsR and BCAM1871, which fine-tune AHL-dependent QS, are shown in dark gray. The second AHL-dependent QS system, CciIR, which is only present in strains of the ET-12 lineage, is shown in light gray. The dashed lines indicate regulation of target genes through the second messenger c-di-GMP. The dotted line indicates contradictory experimental data regarding the autoregulation of cepR2.Citation31,Citation32
Evidence for overlapping QS operons was also obtained from phenotypic investigations. For example, biofilm formation of a cepI rpfFBc double mutant in strain H111 could only be restored to the level of the wild type when the medium was supplemented with both signal molecules.Citation27 This is fully congruent with the finding that optimal expression of BapA, a large secreted protein required for biofilm formation, is dependent on both QS systems.Citation59 Likewise, maximal proteolytic activity was dependent on both signal molecules although BDSF mutants of strains H111 and J2315 were at least partially complemented in the presence of AHLs.Citation45 This effect has been attributed to the decreased amounts of AHLs produced by the BDSF mutant as a consequence of lowered cepI transcription.Citation27 In addition to cepI, cciI was also found to be BDSF-regulated.Citation54 Taken together, the data suggest that the BDSF- and AHL-dependent QS circuitries interact with each other although the molecular mechanism is currently not understood.
The BDSF-dependent QS system is widespread
Like the CepIR system, the RpfFR system is highly conserved within the Bcc.Citation27,Citation52 Furthermore, by using a BDSF biosensor, the production of BDSF (or other cross-reacting cis-2-unsaturated fatty acids) was detectable in all currently described Bcc species (our unpublished data). These results confirm and extend a previous survey that demonstrated the synthesis of BDSF in nine Bcc strains (B. ambifaria, B. anthina, B. cenocepacia, B. dolosa, B. lata, B. multivorans, B. pyrrocinia, B. stabilis, and B. vietnamiensis) by using high-performance liquid chromatography (HPLC) and mass spectrometry.Citation52 This study also showed that B. anthina, B. stabilis, and B. pyrrocinia synthesize, in addition to BDSF, the DSF-family compound cis,cis-11-methyldodeca-2,5-dienoic acid. B. multivorans was found to produce not only these two molecules but also DSF.Citation52 It has been suggested that the synthesis of different DSF family molecules is due to the genetic background of the bacterial strain rather than to variations in the BDSF synthase genes.Citation44,Citation52 These data clearly show that cell-to-cell communication by the aid of cis-2-unsaturated fatty acids is a widespread phenomenon within the Bcc. It is also worth noting that RpfFBc homologs have also been identified in Burkholderia species not belonging to the Bcc, and in other genera including Achromobacter, Yersinia, Serratia, Enterobacter, Pantoea, Cronobacter, Rahnella, Erwinia, and Yokenella.Citation46 This supports the idea that DSF family signal molecules may also serve as interspecies signals.Citation47,Citation49,Citation53
The Role of QS in Pathogenicity of Members of the Bcc
The first Bcc strain was isolated from macerated onion tissue.Citation60 This organism, subsequently named B. cepacia, was found to be the causative agent of soft rot of onions. Other Bcc species have also been described as phytopathogens of banana (B. cenocepacia) and apricot (B. seminalis).Citation61-Citation63 However, their pathogenic potential is not restricted to plants. Bcc species are important opportunistic pathogens and are particularly problematic for individuals suffering from CF, chronic granulomatous disease or in immunocompromised patients. The clinical manifestation of a Bcc infection in CF patients ranges from asymptomatic carriage to a rapidly progressing fatal pneumonia, the so-called “cepacia syndrome”.Citation2 Importantly, all but two Bcc species (B. latens and B. metallica) have been isolated from both environmental and clinical sources and thus it is not possible to distinguish between environmental and clinical strains.Citation2,Citation4,Citation64 For example, from 381 clinical isolates examined in a study by Baldwin et al., more than 20% were indistinguishable by multilocus sequence typing (MLST) from environmental source isolates.Citation65 Using PFGE fingerprinting along with other typing methods, LiPuma et al. showed that the epidemic CF strain prevalent in the mid-Atlantic region of the United States and Europe is identical to isolates recovered from agricultural soil.Citation66 Furthermore, a recent study showed that Bcc species isolated from patient sputum and from the waterbody of a freshwater lake in China showed similar virulence properties in different pathogenicity models.Citation67 Hence, in the absence of patient-to-patient transmission, the natural environment is the most likely source of Bcc infections in humans.Citation6
Over the past decade, substantial progress has been made in identifying and characterizing the virulence determinants and infection mechanisms of Bcc strains (for a review see ref. Citation68). Since Bcc strains can use a wide range of plants and animals as infection hosts, diverse disease models have been developed. Plant models include alfalfa (Medicago sativa) seedlings, onion (Allium cepa), and lettuce (Lactuca sativa L. var longifolia); invertebrate models employ nematodes (Caenorhabditis elegans), the larvae of the greater wax moth (Galleria mellonella) and fruit flies (Drosophila melanogaster); vertebrate infection models make use of zebrafish (Danio rerio) embryos, rats, and mice.Citation67-Citation70 The role of QS in virulence has been investigated for several Bcc strains in various infection models. The results of these investigations have been recently summarized in an excellent review by Subramoni and Sokol.Citation70 The bottom line is that both AHL-dependent QS systems (CepIR and CciIR) contribute to virulence in different vertebrate infection models whereas only the CepIR system is required for virulence in alfalfa (in the case of strain H111 but not strain K56-2) and C. elegans. Interestingly, the AHL-dependent QS systems seem to have no effect on virulence of B. cenocepacia in the G. mellonella and D. melanogaster infection models. Although it has been reported that a cepI mutant of H111 is less virulent in G. mellonella,Citation69 this turned out to be due to a second-site mutation present in this strain (our unpublished results). By contrast, a cepI mutant of the plant growth-promoting B. ambifaria strain showed reduced survival in the D. melanogaster host model.Citation71 Mutants defective in BDSF signaling have so far only been constructed in B. cenocepacia strains J2315 and H111. An rpfFBc mutant of J2315 showed decreased mortality in both a mouse agar bead and a zebrafish infection model as well as reduced killing of G. mellonella.Citation45,Citation51,Citation53 Inactivation of either rpfFBc or rpfR attenuated the virulence of the strain H111 in C. elegans.Citation46 It will be of great interest to investigate whether BDSF-deficient mutants are also attenuated in other infection models and to examine the importance of DSF family-based signaling systems for virulence of other Bcc species.
The use of the above mentioned infection models provided convincing evidence that QS is involved in the pathogenicity of Bcc species. Rather than controlling one universal pathogenicity factor, QS regulates the expression of a variety of virulence factors, which vary in their importance for infections in different host organisms.Citation69 For example, the zinc metalloproteases ZmpA and ZmpB, which are regulated by both AHL and BDSF, were found to be important for the pathogenicity in mammalian hosts and D. melanogaster, whereas they do not contribute to pathogenicity in C. elegans, G. mellonella, or alfalfa.Citation27,Citation69,Citation72 The stringently AHL-regulated protein AidA, is essential for killing of C. elegans, but is not required for virulence in any other infection model.Citation69 A comprehensive list of QS-regulated virulence factors can be found in the review of Subramoni and Sokol.Citation70
A few clinical studies have provided evidence that an intact QS system is important for the pathogenicity of Burkholderia species during infection of CF patients. Analysis of sputum samples revealed that AHLs can be detected in the majority of the samples originating from patients infected with B. cepacia or Pseudomonas aeruginosa, but not Staphylococcus aureus.Citation73 This is in accordance with another study, where AHLs could be extracted from mucopurulent respiratory secretions from 13 cystic fibrosis patients infected with P. aeruginosa and/or Bcc strains.Citation74 Furthermore, McKeon et al. showed that a functional QS system is maintained in B. cenocepacia and B. multivorans during the course of infection in CF patients. Forty-five sequential isolates of these two species isolated over a time course of 2 to 16 y from individuals suffering from CF were analyzed for their ability to produce AHLs. Interestingly, all but one isolate produced AHLs. Further analyses revealed that there is a selective advantage for strains with a functional CepIR system and that mutations in these genes are therefore very rare.Citation75 Additional evidence for an important role of AHL-mediated QS was obtained by analyzing the transcriptome of two clonal variants of B. cenocepacia isolated during long-term colonization of a CF patient.Citation76 In this study, more than 1000 genes were found to be differentially expressed, including cciR and 94 genes previously known to be regulated by CciR. Likewise, DSF signals have been shown to be present in CF sputa and correlated well with patient colonization by Stenotrophomonas maltophilia and/or B. cenocepacia (see above).Citation49
The Role of QS in Biofilm Formation of Members of the Bcc
Evidence that has accumulated over the past decade suggests that QS plays an important role in the development of bacterial biofilms. Cells within the biofilm are embedded in a self-produced extracellular matrix and are in close contact to each other, thus representing a high cell density community. The biofilm matrix may also act as a diffusion barrier for signal molecules, creating an ideal environment for the induction of QS.
The influence of AHL-mediated QS on biofilm formation has been investigated using a quorum-quenching approach, i.e., the enzymatic degradation of AHL signal molecules.Citation77 This survey revealed that the CepIR system controls biofilm formation in the great majority of the Bcc strains tested. A role of AHL signal molecules in biofilm formation was first reported for B. cenocepacia H111. In this study it was shown that inactivation of either cepI or cepR impaired biofilm maturation and resulted in thinner biofilms when compared with the one formed by the wild type.Citation16 Subsequent work showed that the influence of AHL-based QS on biofilm formation of B. cenocepacia K56-2 (harboring two QS systems, cepIR and cciIR), is more complex: while mutations in either cepI, cepR or cciR led to reduced biofilms, mutations in cciI or in cepI and cciI did not affect biofilm formation.Citation26 It was not until 2012 that the QS-regulated factors that link QS and biofilm formation were identified.Citation58 To this end, it was necessary to first unravel the underlying molecular mechanism of biofilm development in the Bcc.
QS-regulated functions affecting biofilm formation in members of the Bcc
The initial step in biofilm development is the adhesion of cells to a surface. This event is often dependent on specialized surface appendages such as fimbriae or flagella, which can act as adhesins.Citation78 By the use of transmission electron microscopy (TEM), five types of pili were identified in Bcc strains: mesh (Msh), filamentous (Fil), spine (Spn), spike (Spk), and cable pili (Cbl).Citation79 Although the cable pilus Cbl has an important role in the adhesion to epithelial cells (and is therefore thought to be of clinical relevance), only a few strains from the Bcc harbor the cblA gene coding for this pilus and not all cblA-positive strains produce the pilus.Citation80 In B. cenocepacia H111, a gene encoding a homolog of the E. coli FimA type I pilus was identified and shown to be controlled by the CepIR system.Citation81 This gene (fimA, BCAL1677) is part of an operon that also contains genes coding for a chaperone–usher secretion apparatus (BCAL1678–1681).Citation58,Citation82 Type I fimbriae are important in some organisms for the adhesion to surfaces;Citation83 however, a fimA mutant of B. cenocepacia H111 was neither defective in biofilm biomass nor in biofilm architecture on abiotic surfaces.Citation58,Citation84 Since additional genes coding for pili are present in the H111 genome, including a cluster encoding a type IV Flp-type pilus, it is plausible that these may compensate for the absence of FimA, at least under the conditions tested. In B. pseudomallei the pilA gene, which codes for a type IV pilus, was demonstrated to be essential for microcolony development but was not required for adherence to human cells in culture.Citation85 In conclusion, the exact role of pili in biofilm development of Burkholderia sp remains to be elucidated.
To identify the genes involved in the late stages of biofilm development, a collection of 5000 random transposon insertion mutants in B. cenocepacia H111 was screened using a microtiter dish-based assay.Citation59 Thirteen mutants that exhibited defects in biofilm formation without being impaired in growth were isolated. Inspection of the biofilms formed by these mutants using confocal laser-scanning microscopy (CLSM) revealed dramatic differences in their morphologies when compared with the one of the parent strain. The genes responsible for the different biofilm architectures encoded several functions, including surface proteins, proteins implicated in the biogenesis and maintenance of the outer membrane and, importantly, regulators and proteins involved in QS.Citation59
One of the genes identified in this screen was bapA, which codes for the biofilm associated protein A (BapA). BapA belongs to a family of large surface proteins that are usually secreted via a type I secretion system and are believed to remain loosely associated with the cell surface.Citation86,Citation87 Several members of this family of proteins have been shown to have a role in biofilm formation in different bacterial species. Examples of these large proteins are LapA and LapF from P. putida KT2440,Citation88 Bap of S. aureus,Citation89 Esp of Enterococcus faecalis,Citation90 LapA from P. fluorescens,Citation86 and BapA of Salmonella enterica.Citation87 Importantly, bapA expression was found to be controlled by the CepIR system in a combined transcriptome and proteome analysis of B. cenocepacia H111 and is to date the only member of the family of large surface proteins whose expression is QS-regulated.Citation58 BapA has been shown to be of crucial importance for biofilm formation on abiotic surfaces, influencing both the architecture and the biomass of the biofilm. The bapA gene is co-transcribed with three genes encoding a type I export machinery. Mutants in this exporter were indistinguishable from a bapA mutant, suggesting that it is involved in the secretion of BapA.Citation58
In the study of Inhülsen et al., a third operon, bclACB (BCAM0184-186 in B. cenocepacia J2315), was identified among the AHL-regulated genes that influenced biofilm morphogenesis.Citation58 This operon encodes for three lectins, which share a PA-IIL-like C-terminal domain. BclB and BclC have additional N-terminal domains. BclA forms homodimers with a strict specificity for oligomannose-type oligosaccharides, present on human glycoproteins.Citation91,Citation92 BclC forms hexamers and has an N-terminal domain displaying a TNF-α-like fold with fucose-binding properties. Since BclC contains two different lectin domains, it is considered to be a super-lectin, with dual carbohydrate specificity.Citation93,Citation94 The third lectin produced by B. cenocepacia, BclB, is the least characterized one but it was recently shown that it may be associated with the bacterial cell surface.Citation58 The biofilms formed by a bclACB mutant strain were found to have an altered architecture, with hollow microcolonies that are not observed in wild-type biofilms. Interestingly, this defect could only be rescued after complementation with an intact bclACB operon, suggesting that the three lectins are not redundant and that all three lectins are needed for biofilm structural development.Citation58
The role of EPS in biofilm formation of Bcc strains
In addition to protein components, EPS is an important constituent of the biofilm matrix, affecting cell attachment and the mechanical stability of the biofilm. EPS is produced by the majority of Bcc species and cepacian, a polysaccharide with a branched heptasaccharide repeating unit, was shown to be particularly widespread among Bcc species.Citation95,Citation96 Cepacian plays a role in biofilm maturation but was not required for the initial steps of biofilm development.Citation10 Two gene clusters, bce-I and bce-II, encode the enzymes necessary for cepacian biosynthesis.Citation96,Citation97 Interestingly, no clear correlation could be established between the ability of 108 Bcc strains to produce EPS or to form biofilms in vitro and the clinical outcome of the infections they caused in different patients.Citation10 In another study an inverse correlation between EPS production and decline of CF lung function by Bcc bacteria has been reported.Citation98 This suggests that non-mucoid isolates are associated with increased disease severity while the mucoid phenotype may be associated with bacterial persistence. In support of this hypothesis it has been shown that an EPS-producing clinical B. cenocepacia isolate was able to inhibit chemotaxis and production of reactive oxygen species (ROS) of neutrophils in vitro, both essential components of innate neutrophil-mediated host defenses.Citation99 Although it has been demonstrated that production of EPS is controlled by an AHL-dependent QS system in plant-associated Burkholderia species,Citation100 there is no evidence that this is also the case in members of the Bcc. A possible mechanism involved in the regulation of cepacian biosynthesis is mediated by the RNA chaperone Hfq, a protein known to regulate target mRNAs by small regulatory non-coding RNAs. In the clinical isolate B. cepacia IST408, the deletion of the hfq gene strongly reduces cepacian production, which could be explained by either a small RNA molecule or by pleiotropic effects caused by the lack of Hfq.Citation101
The role of BDSF and c-di-GMP in biofilm formation of Bcc strains
The widespread bacterial second messenger c-di-GMP has been identified as a key player in the transition of bacteria from the planktonic to the sessile lifestyle. It is generally accepted that high levels of intracellular c-di-GMP promote biofilm formation.Citation57
Fazli et al. demonstrated that overproduction of c-di-GMP by expressing the GGDEF domain protein YedQ from E. coli in B. cenocepacia H111 resulted in the formation of a pellicle at the air-liquid interface in a static liquid culture as well as in wrinkled colony morphology on a solid medium.Citation102 The latter phenotype was exploited to identify the c-di-GMP effector BCAM1349, which was shown to be not only required for these two phenotypes but is also important for virulence in a G. mellonella wax moth larvae infection model.Citation102 In a subsequent study the authors showed that BCAM1349 controls expression of a 12-gene cluster, BCAM1330-BCAM1341, which encodes an EPS of unknown structure.Citation84 This polysaccharide has been shown to provide structural stability to flow-cell grown B. cenocepacia H111 biofilms. In the presence of c-di-GMP, BCAM1349 binds to the promoter region of BCAM1330 and stimulates transcription of the gene cluster.
Recent work has demonstrated that biofilm formation of B. cenocepacia is also controlled by the RpfFR QS system.Citation46,Citation51 Mutants in either rpfFBc or rpfR were found to be severely impaired in biofilm formation.Citation46 In the presence of BDSF, RpfR decreases the cellular level of c-di-GMP, thereby affecting several cellular behaviors including biofilm formation (see above). Among the RpfR-regulated genes we identified bapA, explaining at least in part the effect of the BDSF-dependent QS system on biofilm formation of B. cenocepacia.Citation27 Hence, expression of BapA is subject to control by both QS systems operating in this organism, underpinning the central importance of this large surface protein for biofilm formation by B. cenocepacia.
In a recent study a biofilm model was designed that enables long-term selection for daily adherence to and dispersal from a plastic bead in a test tube.Citation12 In this model, cells must form a biofilm on a plastic bead, which is then used to inoculate another tube containing a fresh bead, so that bacteria must remain adherent during transfer and then disperse to colonize the new bead.Citation12 Adaptive mutations that occurred over a time period of approximately 1050 generations were identified by sequencing DNA from mixed communities (metagenomes), the complete genomes of representative clones, and alleles of 60 alternative clones from multiple time points. Most interestingly, mutations in the rpfR gene were found to be critical for the early evolution of the biofilm community. These mutations occurred either in conserved residues of the GGDEF domain, in the PAS sensor domain, or deleted the entire rpfR gene together with 94 other genes. These data suggest that the BDSF signaling system may play a key role for the transition from the planktonic to the biofilm lifestyle and vice versa.
QS affects resistance of Bcc biofilms
It is well established that Bcc strains growing as a biofilm exhibit a markedly increased resistance to both antibiotics and disinfectants, limiting not only the treatment options for CF patients but also complicating the implementation of effective infection control measures in hospitals.Citation103-Citation106 QS appears to be a highly valuable novel target for treatment of biofilm-related Bcc infections,Citation23 as QS inhibitors were shown to interfere effectively with biofilm formation and detachment of B. multivorans and B. cenocepacia.Citation107-Citation110 Most intriguingly, some QS inhibitors, including baicalin hydrate and cinnamaldehyde, have been shown to improve the success of antibiotic treatment in vitro and in vivo by increasing the susceptibility of bacterial biofilms and reducing virulence during infection, demonstrating the potential of QS blockers as antibacterial and antibiofilm agents.Citation111
Outlook
Members of the Bcc are multilingual talents that master at least two chemical languages. The AHL-dependent CepIR system is fully conserved within the Bcc and more than ten years of research have shown that this regulatory circuit controls biofilm formation, proteolytic activity, motility, antifungal activity, and pathogenicity in the large majority of Bcc strains. More recent work has identified several additional regulators, including other QS systems relying on AHLs, LuxR solos, and transcriptional factors, that finetune CepR-dependent gene expression. The BDSF-dependent cell-to-cell communication system is also involved in the regulation of biofilm formation and virulence. Although recent work has shown that the two QS regulons partly overlap it is presently not known whether the two QS systems operate independently of each other or converge and control the expression or the activity status of a yet to be discovered common regulator, which in turn regulates expression of target genes.Citation27 It will therefore be of great importance to identify the c-di-GMP effector that links BDSF perception through RpfR and target gene expression.
The intracellular second messenger c-di-GMP is believed to play a key role for the lifestyle of bacteria, with high levels being typical for sessile cells during chronic infections and low levels for planktonic cells and acute infections.Citation57,Citation112 Interestingly, B. cenocepacia H111 appears to be an exception to this rule, as inactivation of the BDSF QS system leads to an elevated cellular c-di-GMP level and a concomitant reduction in the strain’s ability to form surface-associated biofilms.Citation27,Citation46 On the other hand, artificially elevated intracellular levels of c-di-GMP were shown to promote a wrinkly colony phenotype, pellicle and biofilm formation.Citation102 The transcriptional regulator BCAM1349 was shown to bind c-di-GMP, thereby activating the production of several biofilm matrix components, including polysaccharides and fimbriae. Under which natural conditions the intracellular c-di-GMP levels are elevated and thus promote the formation of a pellicle rather than a submerged biofilm remains to be elucidated.
| Abbreviations: | ||
| Bcc | = | Burkholderia cepacia complex |
| QS | = | quorum sensing |
| CF | = | cystic fibrosis |
| AHL | = | N-acyl homoserine lactone |
| C8-HSL | = | N-octanoyl-homoserine lactone |
| C6-HSL | = | N-hexanoyl-homoserine lactone |
| BDSF | = | Burkholderia diffusible signal factor |
| DSF | = | diffusible signal factor |
| c-di-GMP | = | bis-(3′-5′) cyclic dimeric guanosine monophosphate |
| EPS | = | exopolysaccharides |
Acknowledgments
Work on quorum sensing and biofilm formation has been supported by the Swiss National Science Foundation (Projects 31003A-122013 and 143773).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 2003; 5:719 - 29; http://dx.doi.org/10.1046/j.1462-2920.2003.00471.x; PMID: 12919407
- Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 2005; 3:144 - 56; http://dx.doi.org/10.1038/nrmicro1085; PMID: 15643431
- Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol 2005; 7:1673 - 85; http://dx.doi.org/10.1111/j.1462-2920.2005.00891.x; PMID: 16232283
- Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, et al. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol 2009; 59:102 - 11; http://dx.doi.org/10.1099/ijs.0.001123-0; PMID: 19126732
- Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E, LiPuma JJ, et al. Burkholderia cenocepacia sp. nov.--a new twist to an old story. Res Microbiol 2003; 154:91 - 6; http://dx.doi.org/10.1016/S0923-2508(03)00026-3; PMID: 12648723
- Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 2008; 104:1539 - 51; http://dx.doi.org/10.1111/j.1365-2672.2007.03706.x; PMID: 18217926
- Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 2010; 16:821 - 30; http://dx.doi.org/10.1111/j.1469-0691.2010.03237.x; PMID: 20880411
- LiPuma JJ, Dasen SE, Nielson DW, Stern RC, Stull TL. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet 1990; 336:1094 - 6; http://dx.doi.org/10.1016/0140-6736(90)92571-X; PMID: 1977981
- Conway BA, Greenberg EP. Quorum-sensing signals and quorum-sensing genes in Burkholderia vietnamiensis.. J Bacteriol 2002; 184:1187 - 91; http://dx.doi.org/10.1128/jb.184.4.1187-1191.2002; PMID: 11807080
- Cunha MV, Sousa SA, Leitão JH, Moreira LM, Videira PA, Sá-Correia I. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J Clin Microbiol 2004; 42:3052 - 8; http://dx.doi.org/10.1128/JCM.42.7.3052-3058.2004; PMID: 15243059
- Häussler S, Lehmann C, Breselge C, Rohde M, Classen M, Tümmler B, et al. Fatal outcome of lung transplantation in cystic fibrosis patients due to small-colony variants of the Burkholderia cepacia complex. Eur J Clin Microbiol Infect Dis 2003; 22:249 - 53; PMID: 12687415
- Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc Natl Acad Sci U S A 2013; 110:E250 - 9; http://dx.doi.org/10.1073/pnas.1207025110; PMID: 23271804
- Dales L, Ferris W, Vandemheen K, Aaron SD. Combination antibiotic susceptibility of biofilm-grown Burkholderia cepacia and Pseudomonas aeruginosa isolated from patients with pulmonary exacerbations of cystic fibrosis. Eur J Clin Microbiol Infect Dis 2009; 28:1275 - 9; http://dx.doi.org/10.1007/s10096-009-0774-9; PMID: 19575248
- Lewenza S, Conway B, Greenberg EP, Sokol PA. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol 1999; 181:748 - 56; PMID: 9922236
- Malott RJ, Baldwin A, Mahenthiralingam E, Sokol PA. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia.. Infect Immun 2005; 73:4982 - 92; http://dx.doi.org/10.1128/IAI.73.8.4982-4992.2005; PMID: 16041013
- Huber B, Riedel K, Hentzer M, Heydorn A, Gotschlich A, Givskov M, et al. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 2001; 147:2517 - 28; PMID: 11535791
- Sokol PA, Sajjan U, Visser MB, Gingues S, Forstner J, Kooi C. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 2003; 149:3649 - 58; http://dx.doi.org/10.1099/mic.0.26540-0; PMID: 14663096
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 2005; 21:319 - 46; http://dx.doi.org/10.1146/annurev.cellbio.21.012704.131001; PMID: 16212498
- Shadel GS, Baldwin TO. Positive autoregulation of the Vibrio fischeri luxR gene. LuxR and autoinducer activate cAMP-catabolite gene activator protein complex-independent and -dependent luxR transcription. J Biol Chem 1992; 267:7696 - 702; PMID: 1373136
- Lutter E, Lewenza S, Dennis JJ, Visser MB, Sokol PA. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect Immun 2001; 69:4661 - 6; http://dx.doi.org/10.1128/IAI.69.7.4661-4666.2001; PMID: 11402012
- Venturi V, Friscina A, Bertani I, Devescovi G, Aguilar C. Quorum sensing in the Burkholderia cepacia complex. Res Microbiol 2004; 155:238 - 44; http://dx.doi.org/10.1016/j.resmic.2004.01.006; PMID: 15142620
- Gotschlich A, Huber B, Geisenberger O, Tögl A, Steidle A, Riedel K, et al. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst Appl Microbiol 2001; 24:1 - 14; http://dx.doi.org/10.1078/0723-2020-00013; PMID: 11403388
- Sokol PA, Malott RJ, Riedel K, Eberl L. Communication systems in the genus Burkholderia: global regulators and targets for novel antipathogenic drugs. Future Microbiol 2007; 2:555 - 63; http://dx.doi.org/10.2217/17460913.2.5.555; PMID: 17927476
- Baldwin A, Sokol PA, Parkhill J, Mahenthiralingam E. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia.. Infect Immun 2004; 72:1537 - 47; http://dx.doi.org/10.1128/IAI.72.3.1537-1547.2004; PMID: 14977960
- Malott RJ, Sokol PA. Expression of the bviIR and cepIR quorum-sensing systems of Burkholderia vietnamiensis.. J Bacteriol 2007; 189:3006 - 16; http://dx.doi.org/10.1128/JB.01544-06; PMID: 17277056
- Tomlin KL, Malott RJ, Ramage G, Storey DG, Sokol PA, Ceri H. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl Environ Microbiol 2005; 71:5208 - 18; http://dx.doi.org/10.1128/AEM.71.9.5208-5218.2005; PMID: 16151106
- Schmid N, Pessi G, Deng Y, Aguilar C, Carlier AL, Grunau A, et al. The AHL- and BDSF-dependent quorum sensing systems control specific and overlapping sets of genes in Burkholderia cenocepacia H111. PLoS One 2012; 7:e49966; http://dx.doi.org/10.1371/journal.pone.0049966; PMID: 23185499
- Subramoni S, Venturi V. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 2009; 155:1377 - 85; http://dx.doi.org/10.1099/mic.0.026849-0; PMID: 19383698
- Hoang HH, Becker A, González JE. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J Bacteriol 2004; 186:5460 - 72; http://dx.doi.org/10.1128/JB.186.16.5460-5472.2004; PMID: 15292148
- Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol 2006; 188:3365 - 70; http://dx.doi.org/10.1128/JB.188.9.3365-3370.2006; PMID: 16621831
- Malott RJ, O’Grady EP, Toller J, Inhülsen S, Eberl L, Sokol PA. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J Bacteriol 2009; 191:2447 - 60; http://dx.doi.org/10.1128/JB.01746-08; PMID: 19201791
- Ryan GT, Wei Y, Winans SC. A LuxR-type repressor of Burkholderia cenocepacia inhibits transcription via antiactivation and is inactivated by its cognate acylhomoserine lactone. Mol Microbiol 2013; 87:94 - 111; http://dx.doi.org/10.1111/mmi.12085; PMID: 23136852
- O’Grady EP, Viteri DF, Sokol PA. A unique regulator contributes to quorum sensing and virulence in Burkholderia cenocepacia.. PLoS One 2012; 7:e37611; http://dx.doi.org/10.1371/journal.pone.0037611; PMID: 22624054
- O’Grady EP, Nguyen DT, Weisskopf L, Eberl L, Sokol PA. The Burkholderia cenocepacia LysR-type transcriptional regulator ShvR influences expression of quorum-sensing, protease, type II secretion, and afc genes. J Bacteriol 2011; 193:163 - 76; http://dx.doi.org/10.1128/JB.00852-10; PMID: 20971902
- O’Grady EP, Viteri DF, Malott RJ, Sokol PA. Reciprocal regulation by the CepIR and CciIR quorum sensing systems in Burkholderia cenocepacia.. BMC Genomics 2009; 10:441; http://dx.doi.org/10.1186/1471-2164-10-441; PMID: 19761612
- Bernier SP, Nguyen DT, Sokol PA. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect Immun 2008; 76:38 - 47; http://dx.doi.org/10.1128/IAI.00874-07; PMID: 17967860
- Aubert DF, O’Grady EP, Hamad MA, Sokol PA, Valvano MA. The Burkholderia cenocepacia sensor kinase hybrid AtsR is a global regulator modulating quorum-sensing signalling. Environ Microbiol 2013; 15:372 - 85; http://dx.doi.org/10.1111/j.1462-2920.2012.02828.x; PMID: 22830644
- Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, et al. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2008; 2:27 - 36; http://dx.doi.org/10.1038/ismej.2007.76; PMID: 18049456
- Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, et al. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol 1997; 24:555 - 66; http://dx.doi.org/10.1046/j.1365-2958.1997.3721736.x; PMID: 9179849
- Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, et al. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol 2004; 51:903 - 12; http://dx.doi.org/10.1046/j.1365-2958.2003.03883.x; PMID: 14731288
- He YW, Zhang LH. Quorum sensing and virulence regulation in Xanthomonas campestris.. FEMS Microbiol Rev 2008; 32:842 - 57; http://dx.doi.org/10.1111/j.1574-6976.2008.00120.x; PMID: 18557946
- Deng Y, Wu J, Tao F, Zhang LH. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev 2011; 111:160 - 73; http://dx.doi.org/10.1021/cr100354f; PMID: 21166386
- Ryan RP, Dow JM. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris.. Virulence 2010; 1:404 - 8; http://dx.doi.org/10.4161/viru.1.5.12704; PMID: 21178479
- Bi H, Christensen QH, Feng Y, Wang H, Cronan JE. The Burkholderia cenocepacia BDSF quorum sensing fatty acid is synthesized by a bifunctional crotonase homologue having both dehydratase and thioesterase activities. Mol Microbiol 2012; 83:840 - 55; http://dx.doi.org/10.1111/j.1365-2958.2012.07968.x; PMID: 22221091
- Deng Y, Boon C, Eberl L, Zhang LH. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J Bacteriol 2009; 191:7270 - 8; http://dx.doi.org/10.1128/JB.00681-09; PMID: 19801414
- Deng Y, Schmid N, Wang C, Wang J, Pessi G, Wu D, et al. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc Natl Acad Sci U S A 2012; 109:15479 - 84; http://dx.doi.org/10.1073/pnas.1205037109; PMID: 22949660
- Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiology 2008; 154:1845 - 58; http://dx.doi.org/10.1099/mic.0.2008/017871-0; PMID: 18599814
- Ryan RP, Dow JM. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol 2011; 19:145 - 52; http://dx.doi.org/10.1016/j.tim.2010.12.003; PMID: 21227698
- Twomey KB, O’Connell OJ, McCarthy Y, Dow JM, O’Toole GA, Plant BJ, et al. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa.. ISME J 2012; 6:939 - 50; http://dx.doi.org/10.1038/ismej.2011.167; PMID: 22134647
- Hall RA, Turner KJ, Chaloupka J, Cottier F, De Sordi L, Sanglard D, et al. The quorum-sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans.. Eukaryot Cell 2011; 10:1034 - 42; http://dx.doi.org/10.1128/EC.05060-11; PMID: 21666074
- Ryan RP, McCarthy Y, Watt SA, Niehaus K, Dow JM. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia.. J Bacteriol 2009; 191:5013 - 9; http://dx.doi.org/10.1128/JB.00473-09; PMID: 19482924
- Deng Y, Wu J, Eberl L, Zhang LH. Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol 2010; 76:4675 - 83; http://dx.doi.org/10.1128/AEM.00480-10; PMID: 20511428
- McCarthy Y, Yang L, Twomey KB, Sass A, Tolker-Nielsen T, Mahenthiralingam E, et al. A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia.. Mol Microbiol 2010; 77:1220 - 36; http://dx.doi.org/10.1111/j.1365-2958.2010.07285.x; PMID: 20624216
- Udine C, Brackman G, Bazzini S, Buroni S, Van Acker H, Pasca MR, et al. Phenotypic and genotypic characterisation of Burkholderia cenocepacia J2315 mutants affected in homoserine lactone and diffusible signal factor-based quorum sensing systems suggests interplay between both types of systems. PLoS One 2013; 8:e55112; http://dx.doi.org/10.1371/journal.pone.0055112; PMID: 23383071
- Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 2011; 65:261 - 86; http://dx.doi.org/10.1146/annurev-micro-121809-151631; PMID: 21663441
- King-Scott J, Konarev PV, Panjikar S, Jordanova R, Svergun DI, Tucker PA. Structural characterization of the multidomain regulatory protein Rv1364c from Mycobacterium tuberculosis.. Structure 2011; 19:56 - 69; http://dx.doi.org/10.1016/j.str.2010.11.010; PMID: 21220116
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 2013; 77:1 - 52; http://dx.doi.org/10.1128/MMBR.00043-12; PMID: 23471616
- Inhülsen S, Aguilar C, Schmid N, Suppiger A, Riedel K, Eberl L. Identification of functions linking quorum sensing with biofilm formation in Burkholderia cenocepacia H111. Microbiologyopen 2012; 1:225 - 42; http://dx.doi.org/10.1002/mbo3.24; PMID: 22950027
- Huber B, Riedel K, Köthe M, Givskov M, Molin S, Eberl L. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol Microbiol 2002; 46:411 - 26; http://dx.doi.org/10.1046/j.1365-2958.2002.03182.x; PMID: 12406218
- Burkholder W. Sour skin, a bacterial rot of onion bulbs. Phytopathology 1950; 64:468 - 75
- Fang Y, Li B, Wang F, Liu B, Wu Z, Su T, et al. Bacterial fruit rot of apricot caused by Burkholderia cepacia in china. Plant Pathol J 2009; 25:429 - 32; http://dx.doi.org/10.5423/PPJ.2009.25.4.429
- Lee Y-A, Chan C-W. Molecular typing and presence of genetic markers among strains of banana finger-tip rot pathogen, Burkholderia cenocepacia, in Taiwan. Phytopathology 2007; 97:195 - 201; http://dx.doi.org/10.1094/PHYTO-97-2-0195; PMID: 18944375
- Li B, Fang Y, Zhang G, Yu R, Lou M, Xie G, et al. Molecular characterization of Burkholderia cepacia complex isolates causing bacterial fruit rot of apricot. Plant Pathol J 2010; 26:223 - 30; http://dx.doi.org/10.5423/PPJ.2010.26.3.223
- Vanlaere E, Lipuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, et al. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol 2008; 58:1580 - 90; http://dx.doi.org/10.1099/ijs.0.65634-0; PMID: 18599699
- Baldwin A, Mahenthiralingam E, Drevinek P, Vandamme P, Govan JR, Waine DJ, et al. Environmental Burkholderia cepacia complex isolates in human infections. Emerg Infect Dis 2007; 13:458 - 61; http://dx.doi.org/10.3201/eid1303.060403; PMID: 17552100
- LiPuma JJ, Spilker T, Coenye T, Gonzalez CF. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 2002; 359:2002 - 3; http://dx.doi.org/10.1016/S0140-6736(02)08836-0; PMID: 12076559
- Ibrahim M, Tang Q, Shi Y, Almoneafy A, Fang Y, Xu L, et al. Diversity of potential pathogenicity and biofilm formation among Burkholderia cepacia complex water, clinical, and agricultural isolates in China. World J Microbiol Biotechnol 2012; 28:2113 - 23; http://dx.doi.org/10.1007/s11274-012-1016-3; PMID: 22806034
- Loutet SA, Valvano MA. A decade of Burkholderia cenocepacia virulence determinant research. Infect Immun 2010; 78:4088 - 100; http://dx.doi.org/10.1128/IAI.00212-10; PMID: 20643851
- Uehlinger S, Schwager S, Bernier SP, Riedel K, Nguyen DT, Sokol PA, et al. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect Immun 2009; 77:4102 - 10; http://dx.doi.org/10.1128/IAI.00398-09; PMID: 19528212
- Subramoni S, Sokol PA. Quorum sensing systems influence Burkholderia cenocepacia virulence. Future Microbiol 2012; 7:1373 - 87; http://dx.doi.org/10.2217/fmb.12.118; PMID: 23231487
- Chapalain A, Vial L, Laprade N, Dekimpe V, Perreault J, Déziel E. Identification of quorum sensing-controlled genes in Burkholderia ambifaria. MicrobiologyOpen 2013:1-17.
- Castonguay-Vanier J, Vial L, Tremblay J, Déziel E. Drosophila melanogaster as a model host for the Burkholderia cepacia complex. PLoS One 2010; 5:e11467; http://dx.doi.org/10.1371/journal.pone.0011467; PMID: 20635002
- Middleton B, Rodgers HC, Cámara M, Knox AJ, Williams P, Hardman A. Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol Lett 2002; 207:1 - 7; http://dx.doi.org/10.1111/j.1574-6968.2002.tb11019.x; PMID: 11886742
- Chambers CE, Visser MB, Schwab U, Sokol PA. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol Lett 2005; 244:297 - 304; http://dx.doi.org/10.1016/j.femsle.2005.01.055; PMID: 15766782
- McKeon SA, Nguyen DT, Viteri DF, Zlosnik JE, Sokol PA. Functional quorum sensing systems are maintained during chronic Burkholderia cepacia complex infections in patients with cystic fibrosis. J Infect Dis 2011; 203:383 - 92; http://dx.doi.org/10.1093/infdis/jiq054; PMID: 21208930
- Mira NP, Madeira A, Moreira AS, Coutinho CP, Sá-Correia I. Genomic expression analysis reveals strategies of Burkholderia cenocepacia to adapt to cystic fibrosis patients’ airways and antimicrobial therapy. PLoS One 2011; 6:e28831; http://dx.doi.org/10.1371/journal.pone.0028831; PMID: 22216120
- Wopperer J, Cardona ST, Huber B, Jacobi CA, Valvano MA, Eberl L. A quorum-quenching approach to investigate the conservation of quorum-sensing-regulated functions within the Burkholderia cepacia complex. Appl Environ Microbiol 2006; 72:1579 - 87; http://dx.doi.org/10.1128/AEM.72.2.1579-1587.2006; PMID: 16461713
- Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol 2000; 182:2675 - 9; http://dx.doi.org/10.1128/JB.182.10.2675-2679.2000; PMID: 10781532
- Goldstein R, Sun L, Jiang RZ, Sajjan U, Forstner JF, Campanelli C. Structurally variant classes of pilus appendage fibers coexpressed from Burkholderia (Pseudomonas) cepacia.. J Bacteriol 1995; 177:1039 - 52; PMID: 7532167
- Sajjan U, Liu L, Lu A, Spilker T, Forstner J, LiPuma JJ. Lack of cable pili expression by cblA-containing Burkholderia cepacia complex. Microbiology 2002; 148:3477 - 84; PMID: 12427939
- Riedel K, Arevalo-Ferro C, Reil G, Görg A, Lottspeich F, Eberl L. Analysis of the quorum-sensing regulon of the opportunistic pathogen Burkholderia cepacia H111 by proteomics. Electrophoresis 2003; 24:740 - 50; http://dx.doi.org/10.1002/elps.200390089; PMID: 12601746
- Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeño-Tárraga AM, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 2009; 191:261 - 77; http://dx.doi.org/10.1128/JB.01230-08; PMID: 18931103
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 1998; 30:285 - 93; http://dx.doi.org/10.1046/j.1365-2958.1998.01061.x; PMID: 9791174
- Fazli M, McCarthy Y, Givskov M, Ryan RP, Tolker-Nielsen T. The exopolysaccharide gene cluster Bcam1330-Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen 2013; 2:105 - 22; http://dx.doi.org/10.1002/mbo3.61; PMID: 23281338
- Boddey JA, Flegg CP, Day CJ, Beacham IR, Peak IR. Temperature-regulated microcolony formation by Burkholderia pseudomallei requires pilA and enhances association with cultured human cells. Infect Immun 2006; 74:5374 - 81; http://dx.doi.org/10.1128/IAI.00569-06; PMID: 16926432
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 2003; 49:905 - 18; http://dx.doi.org/10.1046/j.1365-2958.2003.03615.x; PMID: 12890017
- Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penadés JR, et al. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis.. Mol Microbiol 2005; 58:1322 - 39; http://dx.doi.org/10.1111/j.1365-2958.2005.04907.x; PMID: 16313619
- Martínez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol 2010; 77:549 - 61; http://dx.doi.org/10.1111/j.1365-2958.2010.07249.x; PMID: 20545856
- Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 2001; 183:2888 - 96; http://dx.doi.org/10.1128/JB.183.9.2888-2896.2001; PMID: 11292810
- Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol 2001; 67:4538 - 45; http://dx.doi.org/10.1128/AEM.67.10.4538-4545.2001; PMID: 11571153
- Lameignere E, Malinovská L, Sláviková M, Duchaud E, Mitchell EP, Varrot A, et al. Structural basis for mannose recognition by a lectin from opportunistic bacteria Burkholderia cenocepacia.. Biochem J 2008; 411:307 - 18; http://dx.doi.org/10.1042/BJ20071276; PMID: 18215132
- Marchetti R, Malinovska L, Lameignère E, Adamova L, de Castro C, Cioci G, et al. Burkholderia cenocepacia lectin A binding to heptoses from the bacterial lipopolysaccharide. Glycobiology 2012; 22:1387 - 98; http://dx.doi.org/10.1093/glycob/cws105; PMID: 22763039
- Sulák O, Cioci G, Delia M, Lahmann M, Varrot A, Imberty A, et al. A TNF-like trimeric lectin domain from Burkholderia cenocepacia with specificity for fucosylated human histo-blood group antigens. Structure 2010; 18:59 - 72; http://dx.doi.org/10.1016/j.str.2009.10.021; PMID: 20152153
- Sulák O, Cioci G, Lameignère E, Balloy V, Round A, Gutsche I, et al. Burkholderia cenocepacia BC2L-C is a super lectin with dual specificity and proinflammatory activity. PLoS Pathog 2011; 7:e1002238; http://dx.doi.org/10.1371/journal.ppat.1002238; PMID: 21909279
- Herasimenka Y, Cescutti P, Impallomeni G, Campana S, Taccetti G, Ravenni N, et al. Exopolysaccharides produced by clinical strains belonging to the Burkholderia cepacia complex. J Cyst Fibros 2007; 6:145 - 52; http://dx.doi.org/10.1016/j.jcf.2006.06.004; PMID: 16860003
- Ferreira AS, Leitão JH, Silva IN, Pinheiro PF, Sousa SA, Ramos CG, et al. Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions. Appl Environ Microbiol 2010; 76:441 - 50; http://dx.doi.org/10.1128/AEM.01828-09; PMID: 19948863
- Moreira LM, Videira PA, Sousa SA, Leitão JH, Cunha MV, Sá-Correia I. Identification and physical organization of the gene cluster involved in the biosynthesis of Burkholderia cepacia complex exopolysaccharide. Biochem Biophys Res Commun 2003; 312:323 - 33; http://dx.doi.org/10.1016/j.bbrc.2003.10.118; PMID: 14637140
- Zlosnik JE, Hird TJ, Fraenkel MC, Moreira LM, Henry DA, Speert DP. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J Clin Microbiol 2008; 46:1470 - 3; http://dx.doi.org/10.1128/JCM.02273-07; PMID: 18256220
- Bylund J, Burgess LA, Cescutti P, Ernst RK, Speert DP. Exopolysaccharides from Burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. J Biol Chem 2006; 281:2526 - 32; http://dx.doi.org/10.1074/jbc.M510692200; PMID: 16316987
- Suárez-Moreno ZR, Devescovi G, Myers M, Hallack L, Mendonça-Previato L, Caballero-Mellado J, et al. Commonalities and differences in regulation of N-acyl homoserine lactone quorum sensing in the beneficial plant-associated Burkholderia species cluster. Appl Environ Microbiol 2010; 76:4302 - 17; http://dx.doi.org/10.1128/AEM.03086-09; PMID: 20435760
- Sousa SA, Ramos CG, Moreira LM, Leitão JH. The hfq gene is required for stress resistance and full virulence of Burkholderia cepacia to the nematode Caenorhabditis elegans.. Microbiology 2010; 156:896 - 908; http://dx.doi.org/10.1099/mic.0.035139-0; PMID: 19942656
- Fazli M, O’Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, et al. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia.. Mol Microbiol 2011; 82:327 - 41; http://dx.doi.org/10.1111/j.1365-2958.2011.07814.x; PMID: 21883527
- Peeters E, Nelis HJ, Coenye T. Evaluation of the efficacy of disinfection procedures against Burkholderia cenocepacia biofilms. J Hosp Infect 2008; 70:361 - 8; http://dx.doi.org/10.1016/j.jhin.2008.08.015; PMID: 18977555
- Caraher E, Reynolds G, Murphy P, McClean S, Callaghan M. Comparison of antibiotic susceptibility of Burkholderia cepacia complex organisms when grown planktonically or as biofilm in vitro.. Eur J Clin Microbiol Infect Dis 2007; 26:213 - 6; http://dx.doi.org/10.1007/s10096-007-0256-x; PMID: 17265071
- Peeters E, Nelis HJ, Coenye T. In vitro activity of ceftazidime, ciprofloxacin, meropenem, minocycline, tobramycin and trimethoprim/sulfamethoxazole against planktonic and sessile Burkholderia cepacia complex bacteria. J Antimicrob Chemother 2009; 64:801 - 9; http://dx.doi.org/10.1093/jac/dkp253; PMID: 19633000
- Coenye T, Van Acker H, Peeters E, Sass A, Buroni S, Riccardi G, et al. Molecular mechanisms of chlorhexidine tolerance in Burkholderia cenocepacia biofilms. Antimicrob Agents Chemother 2011; 55:1912 - 9; http://dx.doi.org/10.1128/AAC.01571-10; PMID: 21357299
- Brackman G, Risseeuw M, Celen S, Cos P, Maes L, Nelis HJ, et al. Synthesis and evaluation of the quorum sensing inhibitory effect of substituted triazolyldihydrofuranones. Bioorg Med Chem 2012; 20:4737 - 43; http://dx.doi.org/10.1016/j.bmc.2012.06.009; PMID: 22748377
- Huber B, Eberl L, Feucht W, Polster J. Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z Naturforsch C 2003; 58:879 - 84; PMID: 14713169
- Riedel K, Köthe M, Kramer B, Saeb W, Gotschlich A, Ammendola A, et al. Computer-aided design of agents that inhibit the cep quorum-sensing system of Burkholderia cenocepacia.. Antimicrob Agents Chemother 2006; 50:318 - 23; http://dx.doi.org/10.1128/AAC.50.1.318-323.2006; PMID: 16377703
- Brackman G, Hillaert U, Van Calenbergh S, Nelis HJ, Coenye T. Use of quorum sensing inhibitors to interfere with biofilm formation and development in Burkholderia multivorans and Burkholderia cenocepacia.. Res Microbiol 2009; 160:144 - 51; http://dx.doi.org/10.1016/j.resmic.2008.12.003; PMID: 19146953
- Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo.. Antimicrob Agents Chemother 2011; 55:2655 - 61; http://dx.doi.org/10.1128/AAC.00045-11; PMID: 21422204
- Jonas K, Melefors O, Römling U. Regulation of c-di-GMP metabolism in biofilms. Future Microbiol 2009; 4:341 - 58; http://dx.doi.org/10.2217/fmb.09.7; PMID: 19327118