Abstract
Altered expression of Group A Streptococcus (GAS) virulence factors, including the M protein, can result as a consequence of spontaneous genetic changes that occur during laboratory and animal passage. Occurrence of such secondary mutations during targeted gene deletion could confound the interpretation of effects attributable to the function of the gene being investigated. Contradicting reports on whether the sagA/pel locus regulates the M protein-encoding emm might be due to inconsistent occurrence of mutations unrelated with sagA. This study examined the possibility that altered emm expression observed in association with sagA/pel deletion mutants is artifactual. sagA deletion mutants (MGAS2221ΔsagA) of M1T1 isolate MGAS2221 obtained using liquid broth for GAS growth during the deletion process had diminished emm transcription and no detectable M protein production. In contrast, a ΔsagA mutant of another closely genetically related M1T1 isolate had normal emm expression. The sagB gene does not regulate emm; however, one of three MGAS2221ΔsagB mutants had diminished emm expression. The emm regulator mga was downregulated in these M protein expression-negative strains. These results argue that sagA deletion does not directly cause the downregulation of emm expression. Indeed, two MGAS2221ΔsagA mutants obtained using agar plates for GAS growth during the deletion process both had normal emm expression. We conclude that the sagA/pel locus does not regulate emm expression in the M1T1 lineage and provide a protocol for targeted gene deletion that we find less prone to the generation of mutants exhibiting downregulation in emm expression.
Introduction
Group A Streptococcus (GAS) is a major human pathogen that causes various infections, ranging from uncomplicated infections of the pharynx and skin to life-threatening necrotizing fasciitis and streptococcal toxic shock syndrome. GAS produces many virulence factors to mediate its pathogenesis.Citation1 The M protein is a major surface protein, virulence factor, and protective antigen and is required for the resistance of GAS to phagocytosis by neutrophils.Citation2 Thus, downregulation of emm expression commensurately reduces the capacity of GAS to resist killing by neutrophils and to survive in non-immune blood. The emm gene is positively regulated by the virulence regulator Mga.Citation3 Another important virulence factor is streptolysin S (SLS) that contributes to the resistance of GAS to phagocytic clearance and skin and soft-tissue infections.Citation4-Citation7 SLS is a bacteriocin-like toxin consisting of a posttranslationally modified peptide and activity-essential carrier molecules.Citation8 SLS is responsible for the β-hemolytic activity of GAS colonies grown on blood agar media and also has potent cytolytic activities to leukocytes, platelets, and subcellular organelles. The gene locus for SLS biosynthesis consists of nine genes (sagA to sagI).Citation9 SagA encodes pro-SLS peptide, which is posttranslationally modified to active SLS by SagBCD.Citation8
There are contradicting reports as to whether the sagA locus has a regulatory role in the expression of the emm gene, which encodes the M protein. Betschel et al.Citation10 reported no detrimental effect on M protein production of a transposon insertion at the sagA locus in serotype M1 and M18 GAS. However, Li et al.Citation11 found that a serotype M49 GAS mutant with the insertion of a transposon in the promoter region of the sagA gene had downregulated expression of emm and several other virulence factors and designated the locus as pel for the pleiotropic effect locus. Mangold et al.Citation12 subsequently reported evidence that the sagA/pel locus in the M1 strain SF370 encodes a regulatory RNA. In contrast, Datta et al.Citation13 and Perez et al.Citation14 reported that deletion of the sagA/pel locus in M1 isolates does not cause alteration of virulence gene expression.
GAS can undergo variations in laboratory passages, resulting in variants that usually show reduced expression of M protein and other virulence factors.Citation15,Citation16 It is also known that bacteria frequently accumulate mutations in response to nutritional stress during the stationary phase.Citation17 Various investigations of natural populations of GAS have revealed a high frequency of spontaneous mutations occurring in multiple global regulatory genes, including mga, covRS, and ropB that can mediate pleiotropic effects in GAS secreted and surface virulence factors.Citation18-Citation21 The occurrence of a spurious mutation in one of these global regulators could alter the expression of multiple GAS virulence factors. We present evidence consistent with the hypothesis that the inconsistent finding of downregulation of emm expression in association with targeted sagA/pel deletion may be due to spontaneous mutations or phase variations that arise during laboratory manipulations. We conclude that the sagA/pel locus does not have a role in regulation of emm expression in the M1T1 lineage and have doubts about its capacity to regulate emm expression in GAS of other serotype lineages.
Results
Inconsistent results on effect of sagA/pel deletion in M1T1 GAS isolates on emm expression
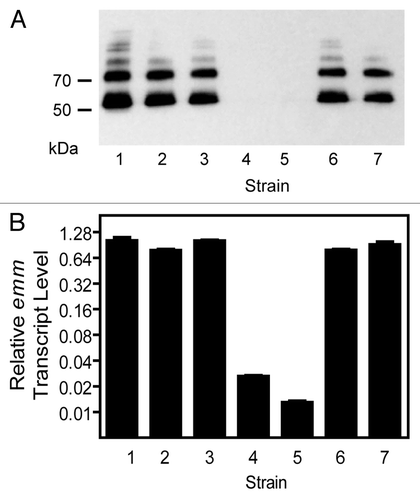
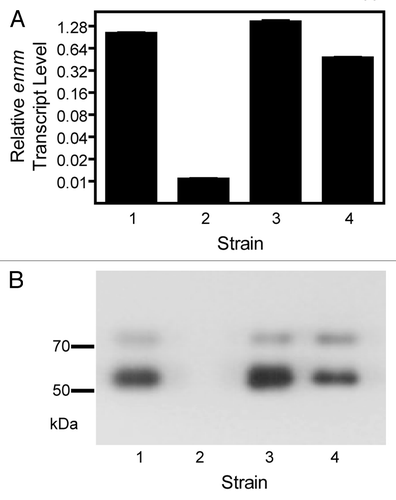
For a separate study we generated a number of gene deletion mutants of two M1T1 isolates MGAS5005 and MGAS2221, including sagA gene deletion mutants. Our antibiotic selection marker-free MGAS2221ΔsagA and other mutants were generated in two steps using a suicide plasmid that contained the chloramphenicol resistance gene and a DNA insert of joint flanking fragments of target genes, as previously described.Citation22 In the first step, the suicide plasmid was integrated into the MGAS2221 chromosome through a single crossover between one of the sagA flanking fragments on the plasmid and its homologous sequence on the chromosome, resulting in a derivative strain that was resistant to chloramphenicol. In the second step, the chloramphenicol-resistant strain was grown in Todd-Hewitt broth supplemented with 2% yeast extract (THY) for at least 8 serial passages to allow for a second crossover between the other sagA flanking fragment on the plasmid and its homologous sequence on the chromosome, yielding sagA deletion mutants (MGAS2221ΔsagA). One passage equaled to the growth from an optical density at 600 nm (OD600) of 0.05 to the late log growth phase with an OD600 of about 0.7 during day and the growth from OD600 of 0.05 to the late stationary growth phase with settled bacteria to the bottom of culture tubes during night (OD600 at the stationary phase in THY was about 0.9). The deletion mutants were screened by PCR analysis and confirmed by the lack of β-hemolytic activity on blood agar plates (). Because we have previously found that some deletion mutants we generate in GAS exhibit reduced M protein production, we now routinely assess the production of the M protein in any GAS mutant that we generate by western blot analysis. The M protein was detected in western blots for MGAS2221 but not for nine MGAS2221ΔsagA mutants from two independent experiments. The western blotting results for two MGAS2221ΔsagA mutants (MGAS2221ΔsagA-1 and MGAS2221ΔsagA-2) are shown in . Real-time reverse transcription PCR analysis found that the levels of the M protein gene (emm) transcript in MGAS2221ΔsagA-1 and MGAS2221ΔsagA-2 were >40-fold lower than that in MGAS2221 (), indicating that the downregulation of the M protein production was at the transcriptional level. These results were surprising to us since the sagA deletion in MGAS2221 did not affect gene expression of MGAS2221 in the study of Perez et al.Citation14 Puzzlingly, a MGAS5005ΔsagA mutant obtained from MGAS5005 using the same procedure had similar M protein production and emm transcription as the parent strain MGAS5005 (). Thus, as in the literature, we obtained conflicting results on the effect of sagA deletion on emm expression.
Figure 1. (A) PCR confirmation of sagA and sagB deletions in MGAS2221 and MGAS5005. Lanes 1 to 7 are PCR products for sagA deletion, and lanes 8 to 11 are PCR products for sagB deletion. (B) Loss of β-hemolytic activity in sagA and sagB deletion mutants. (C) sagA and sagB deletions had no effect on the SpeB protease production measured as the SpeB activity in the supernatant of overnight cultures. Numbers 1 through 11 in all the panels represent the following strains used in the analyses: 1, MGAS5005; 2, MGAS5005ΔsagA; 3, MGAS2221; 4, MGAS2221ΔsagA-1; 5, MGAS2221ΔsagA-2; 6, MGAS2221ΔsagA-3; 7, MGAS2221ΔsagA-4; 8, MGAS2221; 9, MGAS2221ΔsagB-1; 10, MGAS2221ΔsagB-2; and 11, MGAS2221ΔsagB-3.
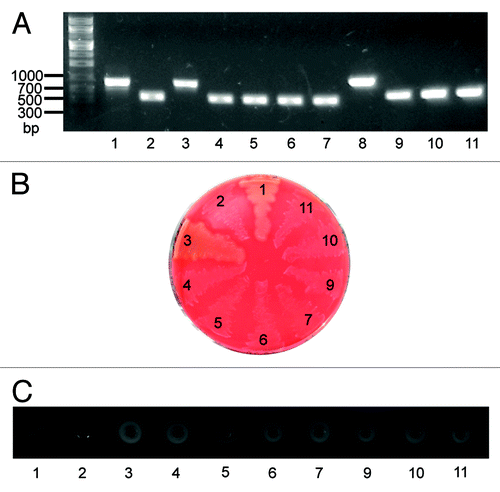
Figure 2. Inconsistent effect of sagA deletion on emm expression in MGAS5005 and MGAS2221. Western blot detecting the production of the M protein (A) and relative levels of emm transcript (B) in the following strains: 1, MGAS5005; 2, MGAS5005ΔsagA; 3, MGAS2221; 4, MGAS2221ΔsagA-1; and 5, MGAS2221ΔsagA-2; 6, MGAS2221ΔsagA-3; 7, MGAS2221ΔsagA-4. Strains 2, 4, and 5 were obtained using THY broth for GAS growth, and strains 6 and 7 were obtained using THY agar plates for GAS growth during the deletion process. Levels of emm mRNA were normalized to that of corresponding wild-type strain.
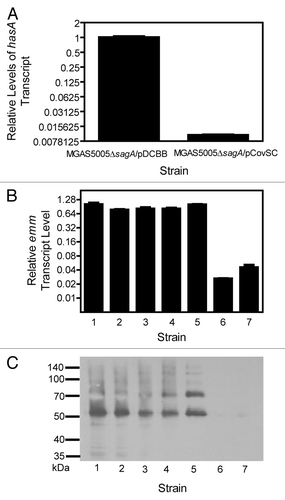
MGAS5005 and MGAS2221 are representative isolates of a prevalent M1T1 clone from an invasive case in Ontario and a scarlet fever patient in Australia, respectively.Citation23 The two isolates have nearly identical genomes, differing by 7 synonymous and 9 non-synonymous single nucleotide polymorphism and an IS element in MGAS5005. In addition, MGAS5005 has a 1-bp deletion at base 83 of the covS gene of the two-component regulatory system CovRS (also known as CsrRS)Citation24 and consequently displays a higher virulence in mouse infection than MGAS2221,Citation25,Citation26 which has the wild-type covS gene. We hypothesized that the difference in effect of sagA deletion on emm expression observed between MGAS2221 and MGAS5005 might be related to their difference in covS alleles. To investigate this possibility, we first transformed MGAS5005ΔsagA with a plasmid that contained the wild-type covS gene (MGAS5005ΔsagA-pCovSC) and deleted the covS gene of MGAS2221ΔsagA-1 (MGAS2221ΔsagAΔcovS). Next, we measured emm transcription and M protein production in MGAS5005ΔsagA-pCovSC and MGAS2221ΔsagAΔcovS. The introduction of the wild-type covS gene in MGAS5005ΔsagA decreased the transcription of the hasA gene by 99% (), indicating that in-trans expression of the wild-type covS gene restored the function of CovRS in MGAS5005ΔsagA. However, the in-trans expression of the functional covS gene did not alter emm expression and M protein production in MGAS5005ΔsagA (). Similarly, emm expression and M protein production were not altered by deletion of covS in MGAS2221ΔsagA-1 (). Thus, difference in effect of sagA deletion on emm expression between MGAS2221 and MGAS5005 was not related to the functionality of the covS gene.
Figure 3. Inconsistent effect of sagA deletion on emm expression was not related to the functionality of the covS gene in MGAS5005 and MGAS2221. (A) Relative levels of hasA mRNA in MGAS5005ΔsagA/pDCBB and MGAS5005ΔsagA/pCovSC. (B and C) Relative levels of emm transcript (A) and western blot detecting the production of the M protein (B) in the following strains: 1, MGAS5005; 2, MGAS5005ΔsagA; 3, MGAS5005ΔsagA/pCovSC; 4, MGAS5005ΔsagA/pDCBB; 5, MGAS2221; 6, MGAS2221ΔsagA-1; and 7, MGAS2221ΔsagAΔcovS. Levels of emm or hasA mRNA were normalized to that of corresponding control strain.
Mixed effects of sagB deletion on emm expression in MGAS2221
The conflicting results obtained for the effect of sagA deletion between MGAS2221 and MGAS5005 suggested that the downregulation of emm expression in MGAS2221ΔsagA-1 and MGAS2221ΔsagA-2 might not be related to deletion of sagA. If altered expression of emm is a common artifact of the targeted gene deletion process and independent of the specific gene being targeted, we should be able to obtain mutants with the downregulation of the M protein production when we delete genes that have been shown to have no role in emm regulation. We tested this hypothesis by deleting the sagB gene because it was shown to not alter emm expression in an investigation of the sagA/pel locus of the M1 SF370 or RND29 strain.Citation12 The procedure used to generate the MGAS2221ΔsagA mutants was followed to delete 395 bp (bases 526-to-920) of the MGAS2221 sagB gene, and three deletion mutants were obtained from a single attempt. One of the three mutants, MGAS2221ΔsagB-1, had downregulated emm transcription and had no detectable M protein production in western blot analysis whereas two of the three MGAS2221 sagB deletion mutants had normal emm transcription and M protein production (). These findings are consistent with our hypothesis that strains exhibiting altered emm expression arise as an artifact of the laboratory manipulations of the deletion process independent of the gene being deleted.
Figure 4. Mixed effects of sagB deletion on emm expression. Relative levels of emm transcript (A) and western blot detecting the production of the M protein (B) in the following strains: 1, MGAS2221; 2, MGAS2221ΔsagB-1; 3, MGAS2221ΔsagB-2; 4, MGAS2221ΔsagB-3. The ΔsagB mutants were obtained using THY broth for GAS growth during the gene deletion process. Levels of emm mRNA were normalized to that of MGAS2221.
The sagA/pel locus in MGAS2221 does not have a role in regulating emm expression
The results on the effect of the sagA and sagB deletions on emm expression imply that the downregulation of emm expression in MGAS2221ΔsagA-1, MGAS2221ΔsagA-2, and MGAS2221ΔsagB-1 was independent of the sagA or sagB deletion. To provide supporting evidence for the contention that sagA does not regulate emm expression, we attempted to generate MGAS2221 sagA deletion mutants with normal emm expression. In our standard GAS gene deletion protocol plasmid integrants resulting from a single crossover homologous recombination were grown in THY broth for many generations to allow for a second crossover event to generate deletion mutants. We speculated that the growth in liquid medium, especially late into the stationary growth phase, may select mutants with downregulation of emm expression. Thus, we grew bacteria on THY agar plates for the second crossover by collecting bacteria from and plating them on agar plates every 12 h. In this way, we obtained two MGAS2221ΔsagA mutants. Both the mutants had wild-type emm transcription and M protein production (lanes or bars 6 and 7 in ). These results strongly argue that the downregulated emm expression in MGAS2221ΔsagA-1 and MGAS2221ΔsagA-2 was not directly caused by the sagA deletion and that the sagA locus has no regulatory role in emm expression.
Downregulation of the M protein production in MGAS2221ΔsagA-1 was stable
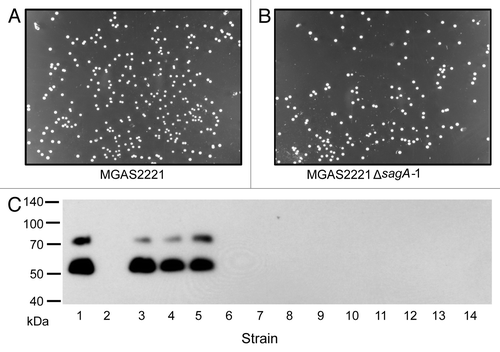
The phase switch variants described in the studies of the Cleary and Podbielski groups have two features.Citation15,Citation16 First, variants show different colony sizes from the starting strains, and, second, the variation is reversible. As shown in , MGAS2221ΔsagA-1 and MGAS2221 had similar colony size. To determine whether the downregulation of the M protein production is stable in the absence of a selection for the restoration of M protein production, MGAS2221ΔsagA-1 was passed on THY agar plate for 8 times, and 9 colonies were randomly chosen and analyzed for the M protein production. All the colonies still had downregulation of the M protein production (lanes 6 to 14 in ). In addition, unlike the phase switch variants in the study of Leonard et al.,Citation16 our deletion mutants with downregulation of the M protein production had no alteration in the SpeB production compared with the strains with normal M protein production (). Taken together these results suggest that our deletion mutants exhibiting downregulated M-protein production differ in character from the phase switching variants described by the Cleary and Podbielski groups.
Figure 5. (A andB) Pictures for colonies of MGAS2221 (A) and its MGAS2221ΔsagA-1 (B) under the reversed background. (C) Western blot for detecting the M protein for the stability of the emm-downregulating phenotype of MGAS2221ΔsagA-1. The mutant and MGAS2221 were passed on THY agar plates for 8 times. After the last passage, GAS bacteria were collected from plates and plated on THY agar plate at appropriate dilution, and 3 (lanes 3–5) and 9 (lanes 6–14) colonies were randomly picked from the MGAS2221 and MGAS2221ΔsagA-1 samples, respectively, for western blotting analyses. Lanes 1 and 2 are the starting MGAS2221 and MGAS2221ΔsagA-1, respectively.
Downregulation of the mga gene in the M protein expression-negative ΔsagA and ΔsagB mutants
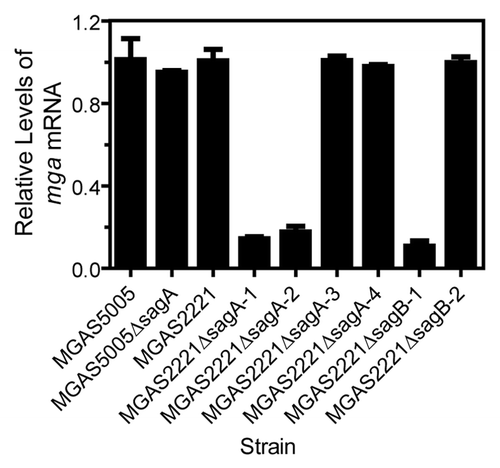
The downregulation of the M protein production in the M protein expression-negative strains might be due to mutations in the promoter region of emm or mga. Thus, we sequenced a 449-bp fragment from upstream base −397 to base 52 in the coding region of mga and a 352-bp fragment covering bases −194 to +158 of emm for the MGAS5005, MGAS5005ΔsagA, MGAS2221, 4 MGAS2221ΔsagA mutants, and 3 MGAS2221ΔsagB strains. All these strains had the identical sequences in these regions. However, levels of the mga transcripts in the M protein expression-negative strains MGAS2221ΔsagA-1, MGAS2221ΔsagA-2, and MGAS2221ΔsagB-1 were 14%, 17%, and 10% of that in MGAS2221, respectively. In contrast, the M protein expression-positive ΔsagA and ΔsagB mutants had similar levels of mga mRNA with the parent strains (). Apparently, the M protein expression-negative phenotype of MGAS2221ΔsagA-1, MGAS2221ΔsagA-2, and MGAS2221ΔsagB-1 was due to the downregulation of the mga regulator. Unfortunately, we do not know what caused the downregulation of mga expression in these strains yet.
Discussion
This report presents findings showing that in the GAS M1T1 genetic lineage sagA deletion is insufficient in and of itself to reproducibly cause downregulation of M protein production. The significance of our findings is 3-fold. First, consistent with the results of Perez et al.,Citation14 our results demonstrate that the sagA locus does not regulate emm expression in the M1T1 lineage. Second, our results raise a concern that targeted gene deletion mutants may have downregulation of M protein production that is not directly associated with the absence of target gene, resulting in a false conclusion on the function of the target gene. Third, we provide a simple protocol to reduce the occurrence of downregulation of M protein production in the generation of GAS gene deletion mutants.
A unique observation in this study is that we obtained mixed results regarding the effect of sagA deletion on emm expression in MGAS2221. Some ΔsagA mutants had diminished emm transcription and non-detectable M protein production whereas others had normal emm expression and M protein production. In other words, the deletion of the sagA gene in a same GAS isolate obtained the results that reflect the contradicting results in a number of published studies regarding a role of the sagA/pel locus in gene regulation.Citation10-Citation14 Our results suggest that the downregulation of emm expression described in the literature could be caused by a separate genetic event during manipulation of GAS for targeted gene inactivation.
The insertion of Tn916 at the promoter region of the sagA gene in a M1 isolate and a M18 strain abolished the β-hemolytic or SLS activity but had no effect on the production of the M protein.Citation10 Sumby and colleagues found that deletion of the sagA/pel locus caused only changes in sagA and sagB expression in M1T1 isolates, including MGAS2221, SF370, and MGAS5005.Citation14 We showed that MGAS2221ΔsagA-3, MGAS2221ΔsagA-4, and MGAS5005ΔsagA mutants had similar levels of emm mRNA with their parent strain according to real-time quantitative RT-PCR. Furthermore, we showed the two major bands of the M protein from the digestion of these ΔsagA and wild-type GAS bacteria with the phage lysin PlyCCitation27 in western blots. The blotting pattern is the same with that in the study of Raz et al.,Citation28 which used the PlyC treatment to release the M protein from the cell wall, and the M protein band with the higher molecular weight was an indication of the M protein linkage to the residual moiety of the peptidoglycan. Thus, the M protein produced in strains of MGAS2221ΔsagA-3, MGAS2221ΔsagA-4, and MGAS5005ΔsagA was full-length and linked to the peptidoglycan. Our data supports the findings of the Sumby studyCitation14 that the sagA/pel locus does not have a regulatory role in gene expression in the M1T1 genetic background and additionally demonstrates that it does not affect posttranscriptional processing and surface localization of intact M protein in M1T1 GAS.
Our results also suggest that the downregulation of the M protein production observed in several previous studies might be artifactual and due to spurious genetic polymorphisms other than changes in the sagA promoter or gene. The insertion of Tn917 transposon at the promoter region of the promoter region of sagA in an M49 serotype GAS strain abolished sagA expression and β-hemolytic activity, and this mutant also displayed diminished or reduced expression of emm, speB, and ska genes.Citation11 Thus, the sagA locus was given the alternate designation, Pel, for pleiotropic effect locus. However, the phenotype of this mutant strain was apparently not stable as passage of this Tn917 insertion mutant in mice selected isolates from the kidney that restored sagA/pel transcription and β-hemolytic activity but did not restore the expression of the other Tn917 insertion-affected virulence genes in the original mutant strain,Citation29 showing that the Tn917 insertion in the upstream of sagA was insufficient in and of itself to cause the pleiotropic effects. Mangold et al. proposed that the sagA locus contains pel as a small regulatory RNA.Citation12 The proposal that the pel locus functions as a regulatory RNA was based on the finding that deletion of sagA/pel (i.e., elimination of transcription was sufficient), but not eliminating sagA/pel translation (i.e., elimination of translation was insufficient), resulted in altered emm and sic transcription relative to the wild-type parent in the serotype M1 strain RDN29. Furthermore, the downregulation of emm expression in the ΔsagA mutant of Mangold et al. could not be repeated in the study of Sumby et al.Citation14 using the strain SF370 (the strain RDN29 is the strain SF370).
Genetic changes responsible for the downregulation of the M protein production in some MGAS2221ΔsagA mutants might be related to the variations that have been termed as phase variation.Citation15,Citation16 It remains unknown whether phase variation is caused by a phase switch. It is possible that genetic changes causing the downregulation of the M protein production were selected in response to environmental stress during stationary growth phase. The viability of GAS decreases dramatically with time at the stationary phase in THY,Citation30 and phase variants are likely selected during stationary phase. Stationary phase mutagenesis is a common phenomenon that accelerates adaption of microbial populations under environmental stress.Citation17 Expression of emm is regulated by a positive regulator Mga, and the Mga regulon is transcribed at high levels in GAS.Citation3,Citation31 The mga expression is downregulated in our M protein expression-negative strains. The downregulation of the Mga regulon could provide an advantage for adaptation of GAS at the stationary phase in vitro under certain condition, which could be the basis for the selection of genetic mutations that downregulate emm transcription during our targeted gene deletion. If this interpretation is valid, such selection pressure would be lessened when GAS grows on THY agar plates.
The downregulation of the M protein production in targeted gene deletion also happened to sagB deletion mutants, suggesting that GAS mutants exhibiting reduced emm expression can be generated by targeted gene deletion independent of the specific gene being targeted. The frequent downregulation of emm expression in targeted gene deletion may be linked to the high frequency of polymorphisms in GAS global regulatory genes shown in numerous publications.Citation18-Citation21 The findings are consistent with the hypothesis that some of the conflicting reports in the sagA/pel literature may be explainable by spontaneous spurious mutations. In addition, independence of frequent downregulation of emm expression on targeted specific gene raises the concern that the phenotype of a specific gene deletion mutant may be caused by a separate genetic alteration that causes downregulation of the M protein production. This situation might have occurred in some previously reported studies. The dltA gene was found to be involved in the resistance of group A Streptococcus (GAS) to destruction by antimicrobial peptide and neutrophils and GAS invasion of epithelial cells through analyzing a dltA deletion mutant;Citation32 however, this same mutant was later found to have diminished expression of the emm gene.Citation33 The emm downregulation in this dltA deletion mutant might be caused by a secondary mutation because a dltA deletion mutant in the study of Cole et al.Citation34 apparently had normal M protein production. Cole et al. found that M protein is essential for in vivo selection of covRS mutations, and their ΔdltA mutant undergoes covRS mutations as its parent strain in in vivo selection. The truncation of the M protein in the sagA insertion mutant in the study of Biswas et al.Citation35 might have been caused by a mutation or deletion in the emm gene. The M protein is a critical GAS virulence factor, and its downregulation due to a secondary mutation could mislead conclusions as to the function of subject gene. Complementation through plasmid-mediated in trans expression is one way to ensure a correct conclusion;Citation23 however, plasmid-based complementation cannot perfectly restore wild-type phenotype sometimes due to the difficulty to express target gene exactly at wild-type level and the loss of the plasmid if experiments are performed in vivo. For the purpose of ruling out a secondary mutation as the cause of a phenotype, the target gene can also be knocked into its deletion mutant without introducing any foreign DNA,Citation22 which would avoid the drawbacks of plasmid-mediated complementation. Besides complementation, it is necessary that GAS mutants in any study should be checked to ensure the normal expression of the emm gene unless a gene deletion is directly linked to emm downregulation. High-throughput DNA sequencing can also be used to assess the information of spontaneous spurious mutations.
Fortunately we found a simple solution to reduce the occurrence of the downregulation of emm expression during targeted gene inactivation in GAS by passing bacteria on THY agar plates. Although the basis for the elimination of the emm downregulation in the plate method is not known, it is practically useful. We have generated more than 30 GAS mutants using this approach, and all of them had normal emm expression. Passing bacteria on agar plates is most likely a useful procedure to reduce the frequency of undesired genetic alterations in targeted gene deletion for other bacteria as well.
Materials and Methods
Bacterial strains and growth
MGAS2221 is a M1T1 GAS strain isolated from a scarlet fever patient, and M1T1 isolate MGAS5005 was isolated from an invasive case in Ontario.Citation23 MGAS5005 and MGAS2221 are very closely genetically related differing by less than 20 single nucleotide polymorphisms; however, the former has a null 1-bp deletion in the covS gene.Citation24 Strains were grown in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY) at 37 °C in a 5% CO2 atmosphere. THY agar was used as the solid media.
Suicide plasmid constructs for antibiotic selection marker-free deletion mutants
In the deletion of the sagA gene, a DNA fragment containing the whole sagA gene and its 24-bp upstream and 164-bp downstream (The downstream sagB was intact and has 62-bp upstream sequence in ΔsagA mutants). Upstream and downstream flanking fragments of this DNA fragment were amplified by PCR using MGAS2221 genomic DNA and primer pairs 5′-TTAGATCTGG TGATGCAAAC TCAATTG-3′/5′-TTCTCGAGGT AACTGATAAG AACGCGAG-3′ and 5′-TTCTCGAGCT AGATAGTACC TGCTAATTAC-3′/5′-TTGGATCCGA AATGTCACGA CACGAAC-3′, respectively. The two PCR fragments were sequentially cloned into suicide vector pGRVCitation30 at the BglII/XhoI and XhoI/BamHI sites, respectively. The fused PCR product was subcloned into the vector pGRV at the BglII and BamHI site, yielding the suicide plasmid pΔsagA.
To obtain a sagB mutant, the upstream (f1)- and downstream (f2)-flanking fragments of an internal 395-bp fragment (bases 526 to 920) of the sagB gene were amplified from the MGAS2221 genomic DNA using the primer pairs 5′-GGGGACAAGT TTGTACAAAA AAGCAGGCTA GCATCTCTAT GTGATAGTG-3′(primer 1)/5′-ATGTCTTGAC ATCAGTAGCA ATGAAGCACA GTTTCTGAGT G-3′ (primer 2) and 5′-TTGCTACTGA TGTCAAGACA TTGACACTCG TAGGAACTAA G-3′ (primer 3)/5′-GGGGACCACT TTGTACAAGA AAGCTGGGTT GATAGAGAGT TGAGTCTTG-3′ (primer 4), respectively. The f1 and f2 PCR fragments were used as template to connect the two fragments in an overlay PCR using primers 1 and 4, and the 21-bp complementary tag sequences that are underlined in primers 2 and 3 bridged the f1 and f2 fragments together in the overlay PCR. The f1–f2 PCR product was cloned into pDONR221 (Life Technologies) using the BP clonase of the Gateway® cloning technology (Life Technology), yielding pDONR221-ΔsagB. The f1–f2 fragment was transferred into pGRV-RFACitation36 in a LR clonase reaction of the Gateway cloning technology, yielding pΔsagB.
Gene deletion procedures that can result in downregulation of emm expression
In the generation of antibiotic selection marker-free gene deletion mutants, pΔsagA or pΔsagB plasmid DNA was introduced into MGAS2221 or MGAS5005 using electroporation, as previously described.Citation30 The suicide plasmid in each experiment was inserted into the GAS chromosome through a homologous recombination event at one flanking fragment of the targeted gene. Bacteria with the first crossover were selected on THY agar plates with 10 mg/l chloramphenicol and contained one copy of full-length target gene and one copy of the shortened target gene with deletion in PCR confirmation analysis. One randomly selected colony, which had the first crossover and normal M protein production, was grown in liquid THY without chloramphenicol for ≥8 serial passages as described earlier. The culture was plated on THY agar plates after the last passage, and resultant colonies were spotted in parallel on THY agar with and without chloramphenicol to identify potential double-crossover strains cured of the suicide plasmid rendering them chloramphenicol-sensitive. These chloramphenicol-sensitive strains were analyzed by PCR to identify deletion mutants that had a shorter PCR product than the wild-type strain.
Gene deletion procedures that reduce the frequency of the downregulation of emm expression
The difference between the procedures that can result in and reduce the frequency of the downregulation of emm expression was in the passage step for the second crossover event. In the procedures to reduce the frequency of the downregulation of emm expression, bacteria from the first crossover step were grown on THY agar plate. At every 12 h after plating, bacteria were recovered from a whole plate, resuspended in 10 ml THY, and vortexed for 2 min. Ten microliters of the vortexed GAS suspension was plated on THY agar plate. Deletion mutants were identified as described above after ≥6 passages.
Complement of the covSΔ1bp pseudo gene in MGAS5005ΔsagA
The plasmid, pCovSC that contained the wild-type covS gene and was used to complement the covSΔ1bp pseudo gene, has been described.Citation23 This plasmid was introduced into MGAS5005ΔsagA by electroporation, and transformants were selected on THY agar plate containing 10 mg chloramphenicol/liter and was referred as MGAS5005ΔsagA-pCovSC. A plasmid vector control strain, MGAS5005ΔsagA-pDCBB, was obtained from the transformation of MGAS5005 with pDCBB.Citation23
Quantitative RT-PCR analysis
Strains were grown at 37 °C, 5% CO2, to an OD600 of 0.2 in THY. Total RNA was isolated from these GAS samples, as described previously.Citation30 High RNA quality was confirmed using an Agilent 2100 Bioanalyzer and an RNA 6000 Lab Chip kit (Agilent Technologies). TaqMan quantitative RT-PCR assays were performed using specific probes and primers () and the ABI 7500 Fast System (Applied Biosystems Inc.), as described previously.Citation37 Control reactions that did not contain reverse transcriptase revealed no contamination of genomic DNA in any RNA sample. All RNA samples were assayed in triplicate, and the levels of emm, hasA, or mga mRNA were compared using the ΔΔCT method with normalization to the mRNA levels of the gyrA gene and presented with normalization to that of corresponding wild-type or control strain.
Table 1. Oligonucleoide primers and fluorescent probes used to quantitate mRNA
Detection of M protein production by western blotting
Wild-type MGAS5005, MGAS2221, and their sagA or sagB deletion mutants in 10 ml cultures were harvested at an OD600 of 0.2 by centrifugation, suspended in 0.2 ml PBS were digested with 10 µg PlyC at room temperature for 1 h. Supernatant samples containing the released cell wall proteins were diluted with 1:60 1× SDS-PAGE loading buffer, and proteins in 10 µl of each sample were resolved by SDS-PAGE and analyzed by western blotting using rabbit anti-M1 protein, as described.Citation22 Briefly, proteins separated by SDS-PAGE were transferred to nitrocellulose membrane (Immobilon-NC, Millipore Corporation) with Towbin transfer buffer using a Trans-Blot Semi-Dry Transfer Cell (Bio-Rad Laboratories) at 15 V for 40 min. The membrane was blocked with 3% bovine serum albumin–0.1% Tween 20 in Tris-buffered saline for 1 h and incubated for 1 h with 1:4000 anti-M1 antisera. The membrane was then rinsed twice and washed three times for 15 min with 0.1% Tween 20 in PBS. The membrane was incubated with goat anti-rabbit IgG (heavy + light chain) HPR conjugate secondary antibodies for 1 h and rinsed and washed as described above. Antigen-antibody reactivity was visualized by enhanced chemiluminescence.
DNA sequencing
A DNA fragment containing the promoter of mga and emm was amplified from testing strains using the paired primers 5′-ATGGTTGTAC CATAACAGTC A-3′/5′-AGTGGAGAGA ACTAAAATTA A-3′ and 5′-CACAACTTAG ACAGCCTAAC T-3′/5′-TCCTAGGGAA GTTATAGAAG A-3′, respectively. DNA sequencing of the amplified PCR products was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit and an Applied Biosystems 3130 genetic analyzer. Sequence data were analyzed using the software Sequencer 5.1 from the Gene Codes Corporation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported in part by grants AI095704, AI097703, and GM103500-09 from the National Institutes of Health, USDA Animal Formula Fund, and the Montana State Agricultural Experimental Station. We thank Dr James Dale at University of Tennessee Health Science Center for providing anti-M protein antisera and Dr Daniel Nelson at University of Maryland for providing PlyC.
References
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 2000; 13:470 - 511; http://dx.doi.org/10.1128/CMR.13.3.470-511.2000; PMID: 10885988
- Whitnack E, Beachey EH. Antiopsonic activity of fibrinogen bound to M protein on the surface of group A streptococci. J Clin Invest 1982; 69:1042 - 5; http://dx.doi.org/10.1172/JCI110508; PMID: 7042754
- Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol 2007; 66:1056 - 65; http://dx.doi.org/10.1111/j.1365-2958.2007.06006.x; PMID: 18001346
- Betschel SD, Borgia SM, Barg NL, Low DE, De Azavedo JC. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Immun 1998; 66:1671 - 9; PMID: 9529097
- Ginsburg I. Is streptolysin S of group A streptococci a virulence factor?. APMIS 1999; 107:1051 - 9; http://dx.doi.org/10.1111/j.1699-0463.1999.tb01509.x; PMID: 10660134
- Engleberg NC, Heath A, Vardaman K, DiRita VJ. Contribution of CsrR-regulated virulence factors to the progress and outcome of murine skin infections by Streptococcus pyogenes. Infect Immun 2004; 72:623 - 8; http://dx.doi.org/10.1128/IAI.72.2.623-628.2004; PMID: 14742501
- Miyoshi-Akiyama T, Takamatsu D, Koyanagi M, Zhao J, Imanishi K, Uchiyama T. Cytocidal effect of Streptococcus pyogenes on mouse neutrophils in vivo and the critical role of streptolysin S. J Infect Dis 2005; 192:107 - 16; http://dx.doi.org/10.1086/430617; PMID: 15942900
- Lee SW, Mitchell DA, Markley AL, Hensler ME, Gonzalez D, Wohlrab A, Dorrestein PC, Nizet V, Dixon JE. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci U S A 2008; 105:5879 - 84; http://dx.doi.org/10.1073/pnas.0801338105; PMID: 18375757
- Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, De Azavedo JC. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun 2000; 68:4245 - 54; http://dx.doi.org/10.1128/IAI.68.7.4245-4254.2000; PMID: 10858242
- Betschel SD, Borgia SM, Barg NL, Low DE, De Azavedo JC. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Immun 1998; 66:1671 - 9; PMID: 9529097
- Li Z, Sledjeski DD, Kreikemeyer B, Podbielski A, Boyle MD. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J Bacteriol 1999; 181:6019 - 27; PMID: 10498714
- Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol Microbiol 2004; 53:1515 - 27; http://dx.doi.org/10.1111/j.1365-2958.2004.04222.x; PMID: 15387826
- Datta V, Myskowski SM, Kwinn LA, Chiem DN, Varki N, Kansal RG, Kotb M, Nizet V. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol 2005; 56:681 - 95; http://dx.doi.org/10.1111/j.1365-2958.2005.04583.x; PMID: 15819624
- Perez N, Treviño J, Liu Z, Ho SC, Babitzke P, Sumby P. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS One 2009; 4:e7668; http://dx.doi.org/10.1371/journal.pone.0007668; PMID: 19888332
- Simpson WJ, Cleary PP. Expression of M type 12 protein by a group A streptococcus exhibits phaselike variation: evidence for coregulation of colony opacity determinants and M protein. Infect Immun 1987; 55:2448 - 55; PMID: 2443452
- Leonard BA, Woischnik M, Podbielski A. Production of stabilized virulence factor-negative variants by group A streptococci during stationary phase. Infect Immun 1998; 66:3841 - 7; PMID: 9673270
- Kivisaar M. Stationary phase mutagenesis: mechanisms that accelerate adaptation of microbial populations under environmental stress. Environ Microbiol 2003; 5:814 - 27; http://dx.doi.org/10.1046/j.1462-2920.2003.00488.x; PMID: 14510835
- Vahling CM, McIver KS. Identification of residues responsible for the defective virulence gene regulator Mga produced by a natural mutant of Streptococcus pyogenes. J Bacteriol 2005; 187:5955 - 66; http://dx.doi.org/10.1128/JB.187.17.5955-5966.2005; PMID: 16109937
- Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog 2010; 6:e1000832; http://dx.doi.org/10.1371/journal.ppat.1000832; PMID: 20368967
- Carroll RK, Shelburne SA 3rd, Olsen RJ, Suber B, Sahasrabhojane P, Kumaraswami M, Beres SB, Shea PR, Flores AR, Musser JM. Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J Clin Invest 2011; 121:1956 - 68; http://dx.doi.org/10.1172/JCI45169; PMID: 21490401
- Horstmann N, Sahasrabhojane P, Suber B, Kumaraswami M, Olsen RJ, Flores A, Musser JM, Brennan RG, Shelburne SA 3rd. Distinct single amino acid replacements in the control of virulence regulator protein differentially impact streptococcal pathogenesis. PLoS Pathog 2011; 7:e1002311; http://dx.doi.org/10.1371/journal.ppat.1002311; PMID: 22028655
- Zhu H, Liu M, Sumby P, Lei B. The secreted esterase of group a streptococcus is important for invasive skin infection and dissemination in mice. Infect Immun 2009; 77:5225 - 32; http://dx.doi.org/10.1128/IAI.00636-09; PMID: 19805529
- Treviño J, Perez N, Ramirez-Peña E, Liu Z, Shelburne SA 3rd, Musser JM, Sumby P. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun 2009; 77:3141 - 9; http://dx.doi.org/10.1128/IAI.01560-08; PMID: 19451242
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2006; 2:e5; http://dx.doi.org/10.1371/journal.ppat.0020005; PMID: 16446783
- Liu M, Zhu H, Li J, Garcia CC, Feng W, Kirpotina LN, Hilmer J, Tavares LP, Layton AW, Quinn MT, et al. Group A Streptococcus secreted esterase hydrolyzes platelet-activating factor to impede neutrophil recruitment and facilitate innate immune evasion. PLoS Pathog 2012; 8:e1002624; http://dx.doi.org/10.1371/journal.ppat.1002624; PMID: 22496650
- Li J, Zhu H, Feng W, Liu M, Song Y, Zhang X, Zhou Y, Bei W, Lei B. Regulation of inhibition of neutrophil infiltration by the two-component regulatory system CovRS in subcutaneous murine infection with group A streptococcus. Infect Immun 2013; 81:974 - 83; http://dx.doi.org/10.1128/IAI.01218-12; PMID: 23319556
- Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci U S A 2006; 103:10765 - 70; http://dx.doi.org/10.1073/pnas.0604521103; PMID: 16818874
- Raz A, Talay SR, Fischetti VA. Cellular aspects of the distinct M protein and SfbI anchoring pathways in Streptococcus pyogenes.. Mol Microbiol 2012; 84:631 - 47; http://dx.doi.org/10.1111/j.1365-2958.2012.08047.x; PMID: 22512736
- Eberhard TH, Sledjeski DD, Boyle MD. Mouse skin passage of a Streptococcus pyogenes Tn917 mutant of sagA/pel restores virulence, beta-hemolysis and sagA/pel expression without altering the position or sequence of the transposon. BMC Microbiol 2001; 1:33; http://dx.doi.org/10.1186/1471-2180-1-33; PMID: 11801184
- Liu M, Hanks TS, Zhang J, McClure MJ, Siemsen DW, Elser JL, Quinn MT, Lei B. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 2006; 152:967 - 78; http://dx.doi.org/10.1099/mic.0.28706-0; PMID: 16549661
- Podbielski A, Woischnik M, Pohl B, Schmidt KH. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med Microbiol Immunol 1996; 185:171 - 81; http://dx.doi.org/10.1007/s004300050028; PMID: 9007823
- Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. D-alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol 2005; 187:6719 - 25; http://dx.doi.org/10.1128/JB.187.19.6719-6725.2005; PMID: 16166534
- Cox KH, Ruiz-Bustos E, Courtney HS, Dale JB, Pence MA, Nizet V, Aziz RK, Gerling I, Price SM, Hasty DL. Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes.. PLoS One 2009; 4:e5366; http://dx.doi.org/10.1371/journal.pone.0005366; PMID: 19401780
- Cole JN, Pence MA, von Köckritz-Blickwede M, Hollands A, Gallo RL, Walker MJ, Nizet V. M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A Streptococcus. MBio 2010; 1:e00191-10; http://dx.doi.org/10.1128/mBio.00191-10; PMID: 20827373
- Biswas I, Germon P, McDade K, Scott JR. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect Immun 2001; 69:7029 - 38; http://dx.doi.org/10.1128/IAI.69.11.7029-7038.2001; PMID: 11598078
- Liu G, Liu M, Xie G, Lei B. Characterization of streptococcal platelet-activating factor acetylhydrolase variants that are involved in innate immune evasion. Infect Immun 2013; 81:3128 - 38; http://dx.doi.org/10.1128/IAI.00398-13; PMID: 23774595
- Lei B, Liu M, Voyich JM, Prater CI, Kala SV, DeLeo FR, Musser JM. Identification and characterization of HtsA, a second heme-binding protein made by Streptococcus pyogenes.. Infect Immun 2003; 71:5962 - 9; http://dx.doi.org/10.1128/IAI.71.10.5962-5969.2003; PMID: 14500516