Abstract
Unique and evolutionarily conserved signaling pathways allow an organism to sense, respond to, and adapt to internal and external environmental cues at its biological niche. In eukaryotic cells, the unfolded protein response (UPR) pathway regulates endoplasmic reticulum (ER) homeostasis upon exposure to environmental changes causing ER stress. The UPR pathway of Cryptococcus neoformans, an opportunistic fungal pathogen, which causes life-threatening meningoencephalitis in immunocompromised individuals, consists of the evolutionarily conserved Ire1 kinase, a unique bZIP transcription factor, Hxl1, and the ER-resident molecular chaperone Kar2/BiP. Although the Cryptococcus UPR pathway regulates ER stress, antifungal drug resistance, and virulence in an Ire1/Hxl1-dependent manner, Ire1 has Hxl1-independent roles in capsule biosynthesis and thermotolerance. In this review, we highlight the conserved and unique features of the Cryptococcus UPR pathway compared with other fungal UPR systems and its importance in the pathogenesis of cryptococcosis and discuss future challenges in this field.
Introduction
The growing elderly population, the extensive use of immunosuppressive agents after organ transplantation or cancer therapy, and the AIDS pandemic has led to an increased number of immunocompromised individuals in past decades. As a result, systemic fungal infection has emerged as a major infectious disease. Life-threatening invasive fungal diseases are mainly caused by opportunistic fungal pathogens such as Candida, Cryptococcus, and Aspergillus.Citation1 With the onset of the AIDS pandemic, the basidiomycetous yeast Cryptococcus has emerged as one of the top ten fatal invasive mycoses, because untreated cryptococcal infection causes lethal meningoencephalitis.
Cryptococcus includes two major pathogenic species, C. neoformans and C. gattii, which have been recently classified as separate species based on distinctive morphological and biochemical characteristics and infection patterns.Citation2,Citation3C. neoformans is the most commonly isolated clade worldwide and mainly infects immunocompromised populations. On the other hand, C. gattii was considered to be geographically restricted to tropical and subtropical regions of the world, but has become more recently isolated from infected immunocompetent individuals in non-tropical regions, such as the Pacific Northwest.Citation4 Nearly 1 000 000 cases of HIV/AIDS-related cryptococcal meningitis occur worldwide every year, causing more than 620 000 deaths.Citation5
Cryptococcus is ubiquitous in environmental niches such as soil, trees, and bird guano. Infectious propagules, in the form of spores or dried yeast cells, are inhaled through the respiratory tract, leading to pulmonary infection. Subsequently, Cryptococcus disseminates from the lung into multiple organs through the bloodstream. This pathogen has a particular tropism to the central nervous system and traverses the blood brain barrier, resulting in meningoencephalitis.Citation6 During the progression of infection, C. neoformans deploys diverse virulence strategies to survive and proliferate in each of the host’s biological niches. Two well characterized virulence factors are the antiphagocytic polysaccharide capsule and the antioxidant melanin.Citation7 Stimulated by several factors such as serum, iron limitation, and physiological CO2 levels,Citation8,Citation9 the capsule is composed of approximately 88% glucuronoxylomannan and 10% galactoxylomannan, and interferes with macrophage phagocytosis or confers direct immunosuppressive activity.Citation10,Citation11 Melanin, which is a brown pigment made of polyphenol complexes, protects cells from environmental UV radiation and oxidative stress in the form of scavenging reactive oxygen species generated by the host defense system during infection. Melanin also enables Cryptococcus to escape from the lung to the central nervous system.Citation12-Citation14
During infection, Cryptococcus experiences dramatic environmental transitions, such as thermal shock, oxidative stress, and high CO2 levels in the host. Therefore, the ability to sense, respond to, and adapt to environmental changes is essential for its survival and proliferation in the host. Cryptococcus exhibits evolutionarily conserved and unique signaling pathways, including HOG (high osmolarity glycerol response), Ras, cAMP/PKA (protein kinase A), Ca2+/calcineurin, and PKC (protein kinase C) pathways, to overcome these external stresses.Citation15-Citation20 In this review, we focus on the conserved and unique features of the Cryptococcus unfolded protein response (UPR), which has recently been shown to play an essential role in endoplasmic reticulum (ER) stress response, in comparison to those of the model yeast, Saccharomyces cerevisiae, and other fungal species. Furthermore, we discuss the potential of the UPR pathway as a novel antifungal therapeutic target and future challenges facing this field.
General Features of UPR Pathways in Yeast and Higher Eukaryotes
The ER is a dynamic organelle with essential roles in protein synthesis, folding, modification, secretion, lipid synthesis, calcium storage and signaling. The accumulation of toxic unfolded or misfolded proteins in the ER triggers “ER stress”, which can also be induced by altered calcium homeostasis and glycosylation, oxidative stress, nutrient starvation, pathogen infection, and activation of inflammation.Citation21 Mounting evidence links ER stress to human diseases as diverse as diabetes, viral infection, Alzheimer disease, cancer, and inflammation.Citation22,Citation23 To mitigate ER stress, eukaryotic cells activate conserved UPR signaling pathways, which regulate the expression of numerous genes encoding ER chaperones and folding enzymes as well as proteins involved in diverse cellular processes.Citation21,Citation24
In mammalian cells, UPR-inducing signals are transduced via three UPR sensors localized at the ER membrane: inositol-requiring protein 1 (IRE1), protein kinase RNA-like ER kinase (PERK), and activating transcription factor 6 (ATF6) ().Citation21 Among these, the IRE1-mediated UPR pathway is the most evolutionarily conserved in eukaryotes. In humans, this pathway consists of IRE1, a type I transmembrane Ser/Thr kinase with an endoribonuclease (RNase) domain, and its downstream transcription factor, XBP1 (X-box binding protein-1). During ER stress, the XBP1 mRNA undergoes unconventional, spliceosome-independent splicing owing to the RNase activity of Ire1. The activated XBP1 transcription factor is subsequently translated from the spliced XBP1 mRNA.Citation21 The unspliced XBP1 mRNA is also translated, but generates a negative regulator of the UPR pathway.Citation25,Citation26
Figure 1. Unfolded protein response (UPR) pathways in eukaryotes. (A) The mammalian UPR pathway consists of three ER-transmembrane sensor proteins, IRE1, PERK, and ATF6. Activation of IRE1 cleaves the 26 nt intron of XBP1u mRNA, and the activated XBP1s bZIP transcription factor upregulates many essential UPR target genes. On the other hand, the translated XBP1u protein appears to sequester XBP1s protein in the cytosol. IRE1 also controls selective mRNA decay (RIDD). Activation of PERK blocks general protein synthesis and increases the specific translation of ATF4 mRNA via phosphorylation of eIF2α. The ATF4 bZIP transcription factor induces expression of UPR target genes. ATF6 is a type II ER transmembrane protein with a bZIP domain. Upon ER stress, ATF6 is translocated to the Golgi and processed proteolytically by site-1 protease (S1P) and site-2 protease (S2P); the ATF6 fragment with the bZIP domain (ATF6f) is then released and translocates to the nucleus to activate UPR genes. (B) The UPR pathway in Arabidopsis thaliana consists of two branches: one involving endoribonuclease IRE1 and the other involving the proteolytic processing of membrane-associated bZIP transcription factors (bZIP17/28). Upon ER stress, IRE1 removes the 23 nt intron of bZIP60 mRNA, resulting in a bZIP protein lacking a transmembrane domain (bZIP60s) via frameshift translation. The bZIP60s transcription factor translocates to the nucleus to activate UPR target genes. Similar to mammalian ATF6, the membrane-associated bZIP transcription factors (bZIP17/28) are processed at the Golgi by S1P and S2P, releasing the truncated versions of bZIP17/28 into the nucleus to activate UPR target genes. Regulated IRE1-dependent decay of specific mRNAs in Arabidopsis has also been observed recently. (C) The yeast Saccharomyces UPR pathway is composed of the Ire1 kinase and the Hac1 bZIP transcription factor. Accumulation of unfolded or misfolded proteins in the ER lumen causes Ire1 to dimerize and trans-autophosphorylate through its kinase domain. The activated Ire1 kinase removes the unconventional intron (252 nt) of the HAC1 mRNA and a tRNA ligase, Rlg1, joins the two exons without the help of conventional spliceosomes. Spliced HAC1 mRNA is translated to produce an active Hac1 protein, which translocates to the nucleus to upregulate expression of UPR target genes encoding ER-resident chaperones and other proteins. “K” and “R” in Ire1 indicate the kinase and ribonuclease domains, respectively.
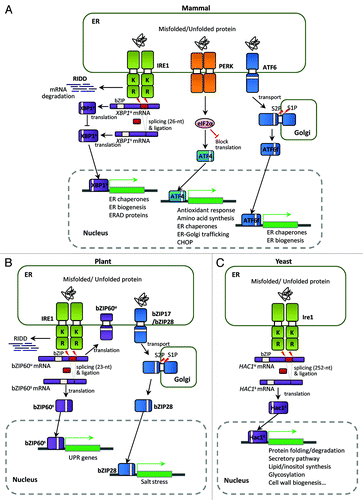
In plants, two branches of the UPR pathway were discovered to play essential roles in ER stress response (). Similar to mammalian ATF6, the ER membrane-associated basic domain/leucine zipper (bZIP) transcription factors bZIP17 and bZIP28 undergo proteolytic activation to regulate ER stress response gene expression.Citation27 Recently, an IRE1-mediated bZIP60 mRNA splicing event was identified in plants. Unspliced bZIP60 mRNA encodes a membrane-associated protein with a bZIP domain and a single transmembrane domain. ER stress stimulates IRE1 to mediate an unconventional splicing event in the bZIP60 mRNA, removing a 23 nt intron to produce a bZIP factor lacking the transmembrane domain but possessing a putative nuclear targeting signal. The resulting bZIP60 protein translocates to the nucleus to activate UPR target genes.Citation28
In S. cerevisiae, a conserved Ire1 sensor and an XBP1 ortholog, Hac1, were discovered and characterized as key components in the yeast UPR pathway ().Citation29 Ire1 senses ER stress in the ER lumen and undergoes autophosphorylation and dimerization for its own activation. Subsequently, activated Ire1 removes an unconventional intron from the HAC1 mRNA, resulting in an active bZIP transcription factor.Citation30 Unlike the human XBP1 mRNA, the unspliced HAC1 mRNA is not translated because of long-range base pairing between the HAC1 5′-untranslated region (5′-UTR) and its intron. The Ire1-dependent unconventional splicing of the HAC1 mRNA is critical for activation of a plethora of UPR target genes.Citation31 Recent studies on the UPR pathways of another ascomycetous yeast, Yarrowia lipolytica, as well as pathogenic fungi such as Candida albicans, Aspergillus fumigatus, and Alternaria brassicicola, revealed that the HAC1 orthologs of these organisms are also subject to unconventional splicing upon ER stress.Citation32-Citation35 Their UPR targets include genes with functions in ER stress, protein secretion, morphological differentiation, and fungal virulence.Citation33-Citation35 However, these orthologs have a shorter unconventional intron (19, 20, or 29 nt) than that of S. cerevisiae (252 nt).
The basic features of the Ire1-dependent UPR signaling pathway appear to be well conserved in most eukaryotes. However, recent reports are revealing unexpected variations, such as the lack of Hac1 homolog in the fission yeast Schizosaccharomyces pombeCitation36,Citation37 and the lack of HAC1 homolog splicing in Candida glabrata.Citation38 Moreover, some protozoans do not possess Ire1 or Hac1/Xbp1 homologs.Citation39,Citation40 Therefore, it seems that the Ire1-dependent UPR pathway has undergone extensive divergence during evolution, particularly in terms of the regulatory mechanism of its downstream bZIP transcription factors.
Core Components of the Cryptococcus UPR Pathway
Cryptococcus has an evolutionarily conserved Ire1 kinase/endonuclease as its sole UPR pathway sensor in the ER and is not likely to contain other UPR sensors such as PERK and ATF6.Citation41 The Cryptococcus Ire1 kinase is highly homologous to the S. cerevisiae Ire1 and to IRE1α and IRE1β in humans. Notably, however, Cryptococcus has a unique bZIP transcription factor encoded by HXL1, which is structurally and phylogenetically distant from yeast Hac1/human XBP1. Like other Hac1/XBP1 orthologs, Hxl1 contains a bZIP domain at the N-terminus (). However, Hxl1 expression does not rescue the ER stress-sensitive phenotypes of the S. cerevisiae hac1∆ mutant, the way that Hac1 orthologs do in other ascomycetes, such as Trichoderma reesei and C. albicans.Citation34,Citation42
Upon ER stress, the endonuclease activity of Ire1 removes an unconventional intron, thereby converting unspliced HXL1 mRNA to spliced HXL1 mRNA.Citation41 Although HXL1u and HXL1s mRNAs encode the same bZIP domain at the N-terminus (60 to 125 aa), the HXL1s mRNA produces the active Hxl1 protein with an extended C-terminal domain (406 aa of Hxl1u to 426 aa of Hxl1i) via frameshift translation of its C-terminal exon region. The Ire1-mediated HXL1 mRNA splicing event is well conserved in different serotypes of Cryptococcus strains, including serotype A C. neoformans var grubii (the H99 strain), serotype D C. neoformans var neoformans (JEC21 and B-3501A strains), and serotype B C. gattii (R265 and WM276 strains).Citation41
The target genes regulated by the UPR pathway play critical roles in counteracting ER stress. Transcriptome profiling analysis of the S. cerevisiae UPR pathway defined the “UPR regulon”, which includes genes involved in the protein secretion and modification pathway as well as ER-resident chaperones.Citation24 As a member of the UPR regulon, an ER-resident molecular chaperone Kar2/BiP ortholog was identified and functionally characterized in C. neoformans.Citation41,Citation43 The expression of C. neoformans KAR2 is tightly regulated in an Ire1- and Hxl1-dependent manner upon ER stress. Kar2 is essential for viability of C. neoformans as it is in S. cerevisiae and C. albicans. Constitutive expression of KAR2 by the strong histone H3 promoter partially restores a subset of Ire1- and Hxl1-dependent phenotypes, including the ER stress response, thermotolerance, and cell wall integrity.Citation43 In summary, Cryptococcus contains both evolutionarily conserved and unique UPR components.
Structural Characteristics of Non-Canonical Introns of HXL1 Orthologs in the Basidiomycota
A recent study by Hooks and Griffiths reported the conserved structure of the non-canonical, spliceosome-independent introns of HAC1/XBP1 homologs in 128 of the 156 eukaryotic species that were examined.Citation36 Interestingly, they could not identify HAC1/XBP1 homologs in several fungal phyla including basidiomycota. Using the bZIP sequence of Hxl1 as query, however, we identified Hxl1 homologs with well conserved non-canonical intron structure in some basidiomycetous fungi, including Trichosporon asahii (A1Q2_03745, e-value 5e-12), Tremella mesenterica (fgeneshTM_kg.7_#_94_#_318_2_CCOI_CCON, 1.2E-15), Tremella fuciformis (Tr21-F09, 3E-08), and Dioszegia cryoxerica (fgenesh1_kg.80_#_88_#_Locus1962v1rpkm301.65, 7.2E-17) (). As in the short intron-containing HAC1/XBP1 orthologs in filamentous fungi and animals, the putative splicing sites “CAG|CAG” and “C(U/C)G|CAG” are observed at both of the putative intron borders in basidiomycetous Hxl1 homolog mRNAs (), and the spliced proteins are expected to be generated by +1 translational frameshifting after removing the unconventional intron. Furthermore, as previously reported in other fungi and animals,Citation32,Citation42,Citation44 the secondary mRNA structures of the unconventional introns of basidiomycetous HXL1 homologs are predicted to form stem-loop structures (), which are probably recognized by Ire1 orthologs.Citation44
Figure 2. Conservation of the putative unconventional splicing sites of HXL1 homologs in basidiomycetes. (A) The unconventional intron sequences of HAC1/XBP1/HXL1 homologous mRNAs are aligned. Flanking exon sequences are denoted by uppercase letters and intron sequences by lowercase letters. Length indicates nucleotide length of the non-conventional intron that can be removed by Ire1. The conserved sequences of 5′- and 3′- splicing junctions are represented by sequence logo and dotted box. (B) Predicted secondary mRNA structures of HXL1 homologs in some basidiomycetous fungi. Putative splicing sites in 5′ and 3′ intron borders are located in the loop regions of stem-loop structures. The putative Ire1-mediated splicing sites and introns are indicated with arrowheads and written in lower case, respectively. Conserved sequences in splicing junctions are indicated by dotted boxes. Alignment and RNA secondary structure prediction were performed with the CLC main benchwork 6.8.4 (CLC bio). Nucleotide sequences were retrieved from the NCBI database and fungal genomics resource at JGICitation61: Cryptococcus neoformans (HXL1, CNAG_06134), Trichosporon asahii (A1Q2_03745), Tremella mesenterica (TREMEDRAFT_57223), Tremella fuciformis (bZIP1, GU723640.1), Dioszegia cryoxerica (fgenesh1_kg.80_#_88_#_Locus1962v1rpkm301.65), Yarrowia lipolytica (HAC1, XM_500811.1), Aspergillus nidulans (hacA, AJ413273), Aspergillus fumigatus (hacA, XM_743634), Trichoderma reesei (hac1, AJ413272), Alternaria brassicicola (HacA, AB01954.1), Candida albicans (HAC1, EF655649), Pichia pastoris (HAC1, FN392319), Saccharomyces cerevisiae (HAC1, NC_001138.5), Homo sapiens (XBP1, NM_005080), Caenorhabditis elegans (xbp-1, AF443190), Arabidopsis thaliana (bZIP60, AY045964).
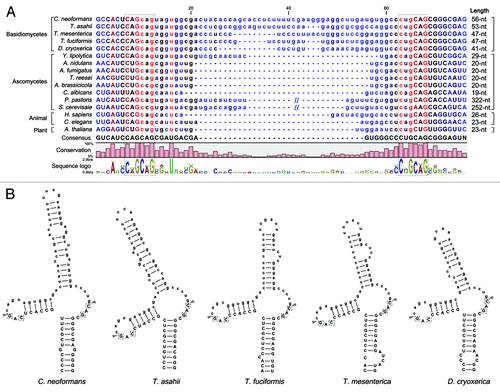
The unconventional introns of basidiomycetous HXL1 homologs (41 to 56 nt) are longer than those of mammalian XBP1 and most ascomycetous HAC1 orthologs (19 to 29 nt), but much shorter than the S. cerevisiae HAC1 intron (252 nt).Citation32-Citation34,Citation42,Citation44,Citation45 In contrast to the S. cerevisiae HAC1 intron, the shorter introns of other fungal HAC1 orthologs and mammalian XBP1 do not contain sequences complementary to their 5′-UTR regions and thus translational attenuation for negative regulation of UPR activation may not occur.Citation31,Citation45 This suggests that there must be other regulatory mechanisms for repressing the unspliced version of HAC1 mRNA (transcriptionally, translationally, or posttranslationally) under normal, unstressed conditions. In mammalian cells, the unspliced XBP1 mRNA is translated under unstressed conditions, but the resulting XBP1 protein is degraded rapidly by the proteasome, thus it is generally undetectable.Citation25 Interestingly, the Cryptococcus HXL1 intron similarly lacks a sequence complementary to any of its 5′-UTR regions, presenting the possibility that unspliced HXL1 mRNA could be translated, but that the Hxl1 protein may be subject to rapid degradation. Another possibility is translational upregulation of Hxl1 as reported in C. albicans, T. reesei, A. nidulans, and A. niger, which express Hac1 mRNAs with truncated 5′-UTRs in response to stress to be translated more efficiently.Citation34,Citation42,Citation46 It will be quite an intriguing endeavor to define the regulatory mechanism of Hxl1 activation and the unique features that distinguish it from other yeast and fungal Hac1 homologs.
Pleiotropic Roles of the UPR Pathway in Cryptococcus
The primary function of the UPR in S. cerevisiae is to relieve ER stress via expression of molecular chaperone genes.Citation24 The Ire1/Hxl1-dependent UPR pathway also promotes resistance to ER stress in the serotype A C. neoformans (the H99 strain) background.Citation41 Both ire1∆ and hxl1∆ mutants are highly susceptible to ER stress agents (e.g., tunicamycin [TM; an N-glycosylation inhibitor] and dithiothreitol [DTT; a reducing agent]). Hxl1 appears to be the only bona fide ER stress response transcription factor downstream of Ire1, since the expression of spliced HXL1 mRNA completely restores wild-type resistance of the ire1∆ mutant to ER and cell wall stresses.Citation41 Similarly, we observed that deletion of IRE1 or HXL1 resulted in increased susceptibility to ER and cell wall stresses in the serotype D C. neoformans and C. gattii R265 (serotype B/C) strain backgrounds (unpublished data by Y.S.B.), indicating that the major roles of the UPR pathway are evolutionarily conserved among pathogenic Cryptococcus species.
In addition to its conserved role in the response to ER stress, the C. neoformans UPR pathway is also involved in resistance to genotoxic stresses, which activate the unconventional splicing of HXL1 mRNA and Ire1/Hxl1-dependent KAR2 induction.Citation43 Supporting this, both ire1∆ and hxl1∆ mutants show increased susceptibility to DNA damaging agents, including hydroxyurea (HU; a ribonucleotide reductase inhibitor) and methyl methane sulfonate (MMS; a DNA alkylating agent that induces DNA base mispairing and blocks replication). DNA damages caused by genotoxic stress are likely to result in production of mutated, abnormal proteins, which may be misfolded or unfolded to trigger ER stress.
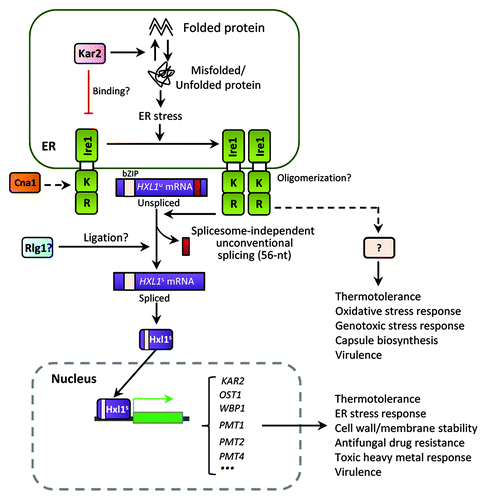
Most notably, the UPR pathway governs the thermotolerance and virulence of Cryptococcus.Citation41 The ability to survive and proliferate at physiological body temperature is an essential virulence factor for most pathogens. Both Ire1 and Hxl1 are required for growth of Cryptococcus at temperatures above 30 °C, and deletion of either gene abolishes its ability to grow at 37 °C. This is likely to be the reason Cryptococcus UPR mutants are avirulent and rapidly cleared during the initial infection stage in the host lung.Citation41 Unlike the case in ER and cell wall stress responses, however, Ire1 and Hxl1 appear to have both redundant and unique roles in thermotolerance based on several observations. First, ire1Δ and hxl1Δ mutants exhibit different levels of temperature sensitivity. In fact, the hxl1∆ mutant is more thermosensitive than the ire1∆ mutant. Second, the expression of spliced HXL1 mRNA only partly restores thermotolerance in the ire1∆ mutant. Third, the ire1Δ mutant harboring the spliced version of HXL1 mRNA is still attenuated in virulence, whereas ire1Δ and hxl1Δ mutants are completely avirulent in a murine model of systemic cryptococcosis. Therefore, Ire1 modulates the thermotolerance and the virulence of C. neoformans in an Hxl1-dependent as well as -independent manner ().
Figure 3. The ER stress response and UPR pathways in C. neoformans. The Cryptococcus UPR pathway consists of the Ire1 kinase, a bZIP transcription factor Hxl1, and their target genes. Upon ER stress, the spliceosome-independent unconventional splicing event in HXL1 mRNA occurs. Activated Hxl1 translocates to the nucleus and induces the expression of UPR target genes such as KAR2, which encodes an ER-resident molecular chaperone. The UPR pathway plays Ire1/Hxl1-dependent roles in ER stress response, antifungal drug resistance, and virulence. However, Ire1 also appears to have Hxl1-independent functions. Crosstalk between the UPR and calcineurin pathways via Cna1 is also indicated in Cryptococcus. Black arrows represent positive regulation or activation whereas red barred lines indicate negative regulation or repression. Dotted arrows indicate potential or unclear regulation.
Hxl1-Independent Ire1 Functions in the Cryptococcus UPR Pathway
Hac1 is the only known Ire1 substrate in S. cerevisiae and no Hac1-independent functions of the UPR pathway have been identified in this yeast model. Although the functions of the UPR pathway are widely conserved in response to ER stress and cell wall stress, Ire1 and its downstream factors, Hac1 in fungi and XBP1 in humans, appear to have distinct roles in response to different environmental cues. Comprehensive gene expression analysis of the UPR pathways in Caenorhabditis elegans and A. fumigatus strongly support the divergent regulation of Ire1 and Hac1 orthologs.Citation47,Citation48 Moreover, in metazoans, the endoribonuclease activity of Ire1 was directly involved in controlling the expression of a subset of genes encoding ER proteins by degrading mRNA to reduce the unfolded protein load in the ER lumen.Citation49,Citation50 It was thus proposed that Ire1 selectively degrades groups of ER-bound mRNAs to relieve the burden of unfolded proteins in the ER lumen via the regulated Ire1-dependent decay (RIDD) pathway.Citation49,Citation50 Furthermore, IRE1α is part of a signaling platform called the UPRosome where several cofactors and adaptor proteins are assembled and function to modulate the kinetics and amplitude of downstream signaling in mammals.Citation51
Notably, Cryptococcus Ire1 also appears to have Hxl1-independent functions. One such function is the regulation of antiphagocytic capsule biosynthesis, as the ire1Δ mutant is defective in capsule biosynthesis. This defect cannot be restored by either KAR2 overexpression or integration of the spliced HXL1 mRNA.Citation41,Citation43 Furthermore, phenotypic analyses revealed that Ire1 generally plays more pleiotropic roles than Hxl1 in response to diverse environmental cues. For example, the ire1∆ mutant, but not the hxl1Δ mutant, shows increased sensitivity to diamide,Citation41 flucytosine, and tert-butyl hydroperoxide (unpublished data by Y.S.B.). In response to heavy metal stress (e.g., CdSO4), the hxl1Δ mutant shows increased resistance, while the ire1Δ mutant shows increased susceptibility (unpublished data by YS Bahn). In response to ER stress and thermal shock, representative UPR target genes, such as KAR2, SEC61 (which regulates translocation of misfolded proteins out of the ER), and DER1 (involved in ER-associated degradation), were shown to be upregulated in an Ire1/Hxl1-dependent manner, whereas expression of PMT1 and PMT4 (protein O-mannosyltransferase) genes appeared to be dependent on Hxl1 only.Citation41 These observations strongly suggest that Ire1 has bifurcated signaling branches (), one of which includes Hxl1 to execute conserved roles of the UPR pathway and the other, which excludes Hxl1. On the other hand, Hxl1 might also have an upstream signaling controller(s), other than Ire1.
Crosstalk between the UPR Pathway and Other Signaling Pathways in Cryptococcus
As reported in UPR-defective mutants of other yeasts and fungi,Citation33,Citation34,Citation52,Citation53 the ire1Δ and hxl1Δ mutants exhibit hypersensitivity to cell wall destabilizing agents, such as Calcofluor white (CFW) and Congo red (CR). Although TM treatment mainly results in defective N-glycosylation, it may also affect cell wall integrity. Indeed, the addition of an osmotic stabilizer (e.g., 1 M sorbitol) restores resistance to TM in the UPR mutants of C. neoformans. Furthermore, TM treatment triggers phosphorylation of the Mpk1 MAPK, a component of the cell wall integrity pathway in C. neoformans.Citation41 Notably, perturbation of the UPR pathway significantly increases Mpk1 phosphorylation levels (under both basal and stress conditions), suggesting that there is direct or indirect crosstalk between the UPR pathway and the Mpk1 MAPK pathway.Citation41
Crosstalk between the UPR and calcineurin pathways is also likely in Cryptococcus. Perturbation of the calcineurin signaling pathway, which is involved in Ca2+ homeostasis, cell wall integrity, thermotolerance, and virulence in C. neoformans, affects HXL1 splicing and KAR2 induction under certain conditions (e.g., high temperature) in C. neoformans.Citation41 It was recently reported that deletion of CNB1, the regulatory B subunit of the calcineurin phosphatase, decreases tolerance to ER stress in Candida glabrata, which is in agreement with the fact that C. neoformans and C. gattii cna1Δ mutants lacking the catalytic subunit of calcineurin show growth defects in response to ER stress.Citation38,Citation41,Citation54 Furthermore, in C. glabrata, KAR2 expression is regulated by the calcineurin pathway, but not by the UPR pathway. This species has lost the canonical Hac1-like transcription factor downstream of Ire1. Therefore, it is highly likely that cellular responses to ER stress involve crosstalk between Ire1 and calcineurin in C. glabrata.Citation38
The UPR pathway may also engage in crosstalk with the mRNA degradation machinery in C. neoformans. Recently, Havel et al.Citation55 demonstrated that ER stress-responsive transcripts are regulated at the post-transcriptional level during adaptation of C. neoformans to host physiological temperature. During ER stress response and host temperature adaptation, the decay rates of ER stress-responsive transcripts, including KAR2, OST2 (a subunit of the ER oligosaccharyltransferase complex), and ALG7 (a lipid-linked N-oligosaccharyltransferase), are lower in cells null for the mRNA deadenylase-encoding CCR4 gene.Citation55 Furthermore, RBP4, which encodes an RNA polymerase II subunit, was shown to be involved in destabilizing the KAR2 transcript during temperature upshift.Citation56 Therefore, the mRNA degradation machinery regulated by Ccr4 and Rpb4 appears to provide an additional level of control to the UPR pathway, contributing to the cellular response to ER stress. In summary, the Ire1/Hxl1-dependent UPR pathway serves as a hub in C. neoformans, interacting with other stress-related signaling pathways directly or indirectly to execute more efficient responses to various environmental cues.
Potential for the UPR Pathway as a Novel Antifungal Therapeutic Target
The need for novel antifungal therapeutic targets and drugs has become urgent due to an increasing incidence of invasive fungal infections, toxic drug side effects, and the emergence of drug-resistant strains.Citation57 Recent studies suggest that the UPR pathway could potentially be exploited as a novel antifungal drug target, since fungal UPR pathways play a critical role in antifungal drug resistance.Citation33,Citation38,Citation41,Citation48
A. fumigatus ireAΔ and hacAΔ mutants exhibit increased susceptibility to azole drugs, such as itraconazole and voriconazole, which function through the inhibition of ergosterol biosynthesis.Citation48 The expression levels of some ergosterol biosynthesis genes, including ERG2, ERG11, ERG24, and ERG3, decrease in both ireAΔ and hacAΔ mutants in A. fumigatus and subsequent ergosterol levels in both stains are reduced. In Cryptococcus, ire1Δ and hxl1Δ mutants also show significantly enhanced susceptibility to azole drugs, including fluconazole, ketoconazole, and itraconazole, although the mechanism appears to be different from that of A. fumigatus based on several observations. First, the expression levels of ERG11 and ERG3 in ire1Δ and hxl1Δ mutants are similar to those of wild type (WT) cells. Furthermore, ire1Δ and hxl1Δ mutants are also slightly more susceptible than WT to amphotericin B, which disrupts the ion homeostasis by binding ergosterol and forming channels in the plasma membrane. Amphotericin B therefore exerts antagonistic effects in combination with azole drugs, supporting the fact that perturbation of the UPR pathway does not affect ergosterol biosynthesis in C. neoformans. Second, azole treatment itself causes ER stress in C. neoformans. Treatment with fluconazole activates the UPR pathway via HXL1 unconventional splicing and induces KAR2 expression in WT cells.Citation43 Accordingly, the overexpression of KAR2 partially suppresses azole sensitivity in ire1Δ and hxl1Δ mutants.
Among UPR pathway components, Hxl1 has several advantages as an attractive therapeutic target for the treatment of cryptococcosis. Hxl1 is conserved in different serotypes of both C. neoformans and C. gattii. The deletion of HXL1 not only abolishes the virulence of C. neoformans var grubii, but also enhances azole susceptibility in C. gattii (R265 strain), C. neoformans var neoformans (JEC21 strain) (unpublished data by YS Bahn), and C. neoformans var grubii.Citation41 This suggests that both mono-therapy (with an Hxl1 inhibitor) and combination therapy with a low dose of azole drugs could be very effective for the treatment of cryptococcosis. Particularly, combination therapy may reduce the hepatotoxicity caused by long-term exposure to high doses of azoles. Moreover, Hxl1 is structurally divergent from the host XBP1 transcription factor, suggesting that an Hxl1-specific inhibitor could be designed to avoid any adverse side effects to the host. Interestingly, recent works have revealed that Ire1 has an allosteric site in its dimer interface that may bind some drugs,Citation58 and small molecules that bind the kinase domain of Ire1 can enhance or reduce its activity.Citation59 Taken together, the core components of the Cryptococcus UPR pathway are potential therapeutic targets for the treatment of cryptococcosis.
Conclusions and Future Perspectives
The functions of the UPR pathway governing the ER stress response and host adaptation are evolutionarily conserved in eukaryotes ranging from yeasts to mammals. In the pathogenic species of Cryptococcus, the UPR pathway has pleiotropic roles in regulating diverse environmental stress responses, the ER stress response, in vitro virulence factor production and in vivo virulence. The fact that UPR signaling components strongly promote antifungal drug resistance suggests that Hxl1, which is a transcription factor that is structurally divergent from the host XBP1, is an ideal target for antifungal drug development. Furthermore, as the first basidiomycetous fungus in which the UPR pathway has been systematically characterized, C. neoformans will serve as an excellent model system to understand the conserved and unique features of the UPR in diverse fungal species.
There are several remaining issues to address in the Cryptococcus UPR pathway. Although the unspliced HXL1 mRNA does not appear to be translated,Citation41 its translational repression mechanism remains an interesting subject to be defined. Considering a lack of any obvious long-range base pairing sequence between 5′ UTR and the unconventional HXL1 intron, the translational control mechanism of the unspliced HXL1 mRNA should be different from that of the yeast HAC1 mRNA. Moreover, it remains unclear to what extent the activation of HXL1 is controlled by Ire1. We observed that some portion of Cryptococcus HXL1 mRNA undergoes splicing even under unstressed conditions in the Ire1-dependent manner.Citation41 Nevertheless, almost a negligible amount of Hxl1 proteins seem to be produced under unstressed conditions. Considering that the Hxl1 protein has a PEST domain, which is involved in degradation,Citation60 it could be speculated that the Hxl1 protein remains unstable due to rapid degradation under unstressed conditions, as is the case in mammalian cells.Citation25 Systematic biochemical characterization of Hxl1 protein stability must be performed to address this issue.
Another issue is the identification of Ire1/Hxl1-, Ire1-, and Hxl1-specific regulons in C. neoformans. To this end, comparative transcriptome analysis of ire1Δ and hxl1Δ mutants under various stress conditions could provide useful information. Particularly, investigation of the RNA stability of Ire1-specific target genes would provide insight on the possible regulatory mechanism of Ire1 independent of Hxl1. Recently, studies in S. pombe and C. glabrata revealed that Ire1 degrades a subset of ER-localized mRNAs to relieve ER stress.Citation37,Citation38 We also observed that upon ER stress or thermal shock, a subset of genes show increased mRNA levels in the ire1∆ mutant, but not in the hxl1∆ mutant, compared with the WT C. neoformans.Citation41 Thus, it could be speculated that Hxl1-indepent roles of Ire1 may be partly mediated by the mRNA-decay machinery in C. neoformans.
Finally, it will be also important to elucidate the UPR pathway in C. gattii, which causes fatal disease even in immunocompetent individuals. The UPR mutants of C. neoformans var grubii are completely cleared at the initial stage of infection (within the lung) in an immunodeficient murine model of systemic cryptococcosis (A/Jcr mice).Citation41 Therefore, it will be interesting to examine whether the C. gattii UPR mutants could be similarly cleared in both immunocompetent and immunocompromised murine models. Such comparative analyses of C. gattii and C. neoformans UPR pathways should help us understand the differential pathogenic mechanisms of the two sibling species.
| Abbreviations: | ||
| ATF6 | = | activating transcription factor 6 |
| bZIP | = | basic domain/leucine zipper |
| DTT | = | dithiothreitol |
| ER | = | endoplasmic reticulum |
| HOG | = | high osmolarity glycerol response |
| HU | = | hydroxyurea |
| IRE1 | = | inositol-requiring protein 1 |
| MMS | = | methyl methane sulfonate |
| PERK | = | protein kinase RNA-like ER kinase |
| PKA | = | protein kinase A |
| PKC | = | protein kinase C |
| RIDD | = | regulated Ire1-dependent decay |
| TM | = | tunicamycin |
| UPR | = | unfolded protein response |
| UTR | = | untranslated region |
| XBP1 | = | X-box binding protein-1 |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grants, 2008-0061963, 2010-0024569, 2010-0029117, and 2012-0001150, funded by the Korea government (MEST).
References
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med 2012; 4:65rv13; http://dx.doi.org/10.1126/scitranslmed.3004404; PMID: 23253612
- Sorrell TC. Cryptococcus neoformans variety gattii.. Med Mycol 2001; 39:155 - 68; PMID: 11346263
- Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon 2002; 51:804 - 6; http://dx.doi.org/10.2307/1555045
- Byrnes EJ 3rd, Bartlett KH, Perfect JR, Heitman J. Cryptococcus gattii: an emerging fungal pathogen infecting humans and animals. Microbes Infect 2011; 13:895 - 907; http://dx.doi.org/10.1016/j.micinf.2011.05.009; PMID: 21684347
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 2009; 23:525 - 30; http://dx.doi.org/10.1097/QAD.0b013e328322ffac; PMID: 19182676
- Sukroongreung S, Kitiniyom K, Nilakul C, Tantimavanich S. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans.. Med Mycol 1998; 36:419 - 24; PMID: 10206753
- Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans.. Nat Rev Microbiol 2005; 3:753 - 64; http://dx.doi.org/10.1038/nrmicro1245; PMID: 16132036
- Vartivarian SE, Anaissie EJ, Cowart RE, Sprigg HA, Tingler MJ, Jacobson ES. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis 1993; 167:186 - 90; http://dx.doi.org/10.1093/infdis/167.1.186; PMID: 8418165
- Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest 1985; 76:508 - 16; http://dx.doi.org/10.1172/JCI112000; PMID: 3928681
- Aksenov SI, Babyeva IP, Golubev VI. On the mechanism of adaptation of micro-organisms to conditions of extreme low humidity. Life Sci Space Res 1973; 11:55 - 61; PMID: 12523380
- Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. A yeast under cover: the capsule of Cryptococcus neoformans.. Eukaryot Cell 2003; 2:655 - 63; http://dx.doi.org/10.1128/EC.2.4.655-663.2003; PMID: 12912884
- Nosanchuk JD, Rosas AL, Lee SC, Casadevall A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 2000; 355:2049 - 50; http://dx.doi.org/10.1016/S0140-6736(00)02356-4; PMID: 10885360
- Casadevall A, Rosas AL, Nosanchuk JD. Melanin and virulence in Cryptococcus neoformans.. Curr Opin Microbiol 2000; 3:354 - 8; http://dx.doi.org/10.1016/S1369-5274(00)00103-X; PMID: 10972493
- Zhu X, Williamson PR. Role of laccase in the biology and virulence of Cryptococcus neoformans.. FEMS Yeast Res 2004; 5:1 - 10; http://dx.doi.org/10.1016/j.femsyr.2004.04.004; PMID: 15381117
- Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans.. Mol Biol Cell 2005; 16:2285 - 300; http://dx.doi.org/10.1091/mbc.E04-11-0987; PMID: 15728721
- Bahn YS, Geunes-Boyer S, Heitman J. Ssk2 mitogen-activated protein kinase kinase kinase governs divergent patterns of the stress-activated Hog1 signaling pathway in Cryptococcus neoformans.. Eukaryot Cell 2007; 6:2278 - 89; http://dx.doi.org/10.1128/EC.00349-07; PMID: 17951522
- Kronstad JW, Hu G, Choi J. The cAMP/protein kinase A pathway and virulence in Cryptococcus neoformans.. Mycobiology 2011; 39:143 - 50; http://dx.doi.org/10.5941/MYCO.2011.39.3.143; PMID: 22783095
- Alspaugh JA, Perfect JR, Heitman J. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans.. Fungal Genet Biol 1998; 25:1 - 14; http://dx.doi.org/10.1006/fgbi.1998.1079; PMID: 9806801
- Kraus PR, Heitman J. Coping with stress: calmodulin and calcineurin in model and pathogenic fungi. Biochem Biophys Res Commun 2003; 311:1151 - 7; http://dx.doi.org/10.1016/S0006-291X(03)01528-6; PMID: 14623301
- Kraus PR, Nichols CB, Heitman J. Calcium- and calcineurin-independent roles for calmodulin in Cryptococcus neoformans morphogenesis and high-temperature growth. Eukaryot Cell 2005; 4:1079 - 87; http://dx.doi.org/10.1128/EC.4.6.1079-1087.2005; PMID: 15947200
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8:519 - 29; http://dx.doi.org/10.1038/nrm2199; PMID: 17565364
- Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol 2008; 3:399 - 425; http://dx.doi.org/10.1146/annurev.pathmechdis.3.121806.151434; PMID: 18039139
- Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature 2008; 454:455 - 62; http://dx.doi.org/10.1038/nature07203; PMID: 18650916
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000; 101:249 - 58; http://dx.doi.org/10.1016/S0092-8674(00)80835-1; PMID: 10847680
- Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J Biol Chem 2006; 281:5852 - 60; http://dx.doi.org/10.1074/jbc.M509061200; PMID: 16332684
- Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol 2006; 172:565 - 75; http://dx.doi.org/10.1083/jcb.200508145; PMID: 16461360
- Liu J-X, Howell SH. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 2010; 22:2930 - 42; http://dx.doi.org/10.1105/tpc.110.078154; PMID: 20876830
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci U S A 2011; 108:7247 - 52; http://dx.doi.org/10.1073/pnas.1102117108; PMID: 21482766
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 1996; 87:391 - 404; http://dx.doi.org/10.1016/S0092-8674(00)81360-4; PMID: 8898193
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997; 90:1031 - 9; http://dx.doi.org/10.1016/S0092-8674(00)80369-4; PMID: 9323131
- Rüegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 2001; 107:103 - 14; http://dx.doi.org/10.1016/S0092-8674(01)00505-0; PMID: 11595189
- Oh MH, Cheon SA, Kang HA, Kim JY. Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 2010; 27:443 - 52; http://dx.doi.org/10.1002/yea.1762; PMID: 20162530
- Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, White S, McCarthy JW, Latgé JP, Feldmesser M, et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus.. PLoS Pathog 2009; 5:e1000258; http://dx.doi.org/10.1371/journal.ppat.1000258; PMID: 19132084
- Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, Yin Z, Brown AJ, Archer DB. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans.. Fungal Genet Biol 2008; 45:1235 - 47; http://dx.doi.org/10.1016/j.fgb.2008.06.001; PMID: 18602013
- Joubert A, Simoneau P, Campion C, Bataillé-Simoneau N, Iacomi-Vasilescu B, Poupard P, François JM, Georgeault S, Sellier E, Guillemette T. Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola.. Mol Microbiol 2011; 79:1305 - 24; http://dx.doi.org/10.1111/j.1365-2958.2010.07522.x; PMID: 21251090
- Hooks KB, Griffiths-Jones S. Conserved RNA structures in the non-canonical Hac1/Xbp1 intron. RNA Biol 2011; 8:552 - 6; http://dx.doi.org/10.4161/rna.8.4.15396; PMID: 21593604
- Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, Aragón T, Li H, Walter P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife 2012; 1:e00048; http://dx.doi.org/10.7554/eLife.00048; PMID: 23066505
- Miyazaki T, Nakayama H, Nagayoshi Y, Kakeya H, Kohno S. Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata.. PLoS Pathog 2013; 9:e1003160; http://dx.doi.org/10.1371/journal.ppat.1003160; PMID: 23382685
- Hollien J. Evolution of the unfolded protein response. Biochim Biophys Acta 2013; 1833:2458 - 63; http://dx.doi.org/10.1016/j.bbamcr.2013.01.016; PMID: 23369734
- Joyce BR, Tampaki Z, Kim K, Wek RC, Sullivan WJ Jr.. The unfolded protein response in the protozoan parasite Toxoplasma gondii features translational and transcriptional control. Eukaryot Cell 2013; 12:979 - 89; http://dx.doi.org/10.1128/EC.00021-13; PMID: 23666622
- Cheon SA, Jung KW, Chen YL, Heitman J, Bahn YS, Kang HA. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans.. PLoS Pathog 2011; 7:e1002177; http://dx.doi.org/10.1371/journal.ppat.1002177; PMID: 21852949
- Saloheimo M, Valkonen M, Penttilä M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol Microbiol 2003; 47:1149 - 61; http://dx.doi.org/10.1046/j.1365-2958.2003.03363.x; PMID: 12581366
- Jung KW, Kang HA, Bahn YS. Essential roles of the Kar2/BiP molecular chaperone downstream of the UPR pathway in Cryptococcus neoformans.. PLoS One 2013; 8:e58956; http://dx.doi.org/10.1371/journal.pone.0058956; PMID: 23484059
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002; 415:92 - 6; http://dx.doi.org/10.1038/415092a; PMID: 11780124
- Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 2006; 22:487 - 508; http://dx.doi.org/10.1146/annurev.cellbio.21.122303.120200; PMID: 16822172
- Mulder HJ, Saloheimo M, Penttilä M, Madrid SM. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol Genet Genomics 2004; 271:130 - 40; http://dx.doi.org/10.1007/s00438-003-0965-5; PMID: 14730445
- Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans.. PLoS Genet 2005; 1:e37; http://dx.doi.org/10.1371/journal.pgen.0010037; PMID: 16184190
- Feng X, Krishnan K, Richie DL, Aimanianda V, Hartl L, Grahl N, Powers-Fletcher MV, Zhang M, Fuller KK, Nierman WC, et al. HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus.. PLoS Pathog 2011; 7:e1002330; http://dx.doi.org/10.1371/journal.ppat.1002330; PMID: 22028661
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 2009; 186:323 - 31; http://dx.doi.org/10.1083/jcb.200903014; PMID: 19651891
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006; 313:104 - 7; http://dx.doi.org/10.1126/science.1129631; PMID: 16825573
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012; 13:89 - 102; PMID: 22251901
- Scrimale T, Didone L, de Mesy Bentley KL, Krysan DJ. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae.. Mol Biol Cell 2009; 20:164 - 75; http://dx.doi.org/10.1091/mbc.E08-08-0809; PMID: 18971375
- Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. An extensive circuitry for cell wall regulation in Candida albicans.. PLoS Pathog 2010; 6:e1000752; http://dx.doi.org/10.1371/journal.ppat.1000752; PMID: 20140194
- Chen YL, Lehman VN, Lewit Y, Averette AF, Heitman J. Calcineurin governs thermotolerance and virulence of Cryptococcus gattii. G3 (Bethesda) 2013; 3:527 - 39; http://dx.doi.org/10.1534/g3.112.004242; PMID: 23450261
- Havel VE, Wool NK, Ayad D, Downey KM, Wilson CF, Larsen P, Djordjevic JT, Panepinto JC. Ccr4 promotes resolution of the endoplasmic reticulum stress response during host temperature adaptation in Cryptococcus neoformans.. Eukaryot Cell 2011; 10:895 - 901; http://dx.doi.org/10.1128/EC.00006-11; PMID: 21602483
- Bloom AL, Solomons JT, Havel VE, Panepinto JC. Uncoupling of mRNA synthesis and degradation impairs adaptation to host temperature in Cryptococcus neoformans.. Mol Microbiol 2013; 89:65 - 83; http://dx.doi.org/10.1111/mmi.12258; PMID: 23659661
- Kathiravan MK, Salake AB, Chothe AS, Dudhe PB, Watode RP, Mukta MS, Gadhwe S. The biology and chemistry of antifungal agents: a review. Bioorg Med Chem 2012; 20:5678 - 98; http://dx.doi.org/10.1016/j.bmc.2012.04.045; PMID: 22902032
- Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, Sicheri F, Ron D. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell 2010; 38:291 - 304; http://dx.doi.org/10.1016/j.molcel.2010.04.001; PMID: 20417606
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Stroud RM, Zhang C, Shokat KM, Walter P. Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1. BMC Biol 2011; 9:48; http://dx.doi.org/10.1186/1741-7007-9-48; PMID: 21729334
- Pal B, Chan NC, Helfenbaum L, Tan K, Tansey WP, Gething MJ. SCFCdc4-mediated degradation of the Hac1p transcription factor regulates the unfolded protein response in Saccharomyces cerevisiae.. Mol Biol Cell 2007; 18:426 - 40; http://dx.doi.org/10.1091/mbc.E06-04-0304; PMID: 17108329
- Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 2012; 40:Database issue D26 - 32; http://dx.doi.org/10.1093/nar/gkr947; PMID: 22110030
