Stenotrophomonas maltophilia, which is still defined as an organism of limited pathogenicity, has risen prominently as a nosocomial pathogen. Despite the increase in the spectrum of clinical syndromes associated with S. maltophilia, very little is known about the extracellular enzymes profile, pigment production and motility patterns which may have potential roles in pathogenesis. In this study, we screened and compared an array of extracellular enzymes in S. maltophilia collected from invasive and non-invasive clinical specimens by substrate plate assays. We also grouped the isolates as device related and non-device related and compared the enzyme profile. Our study showed all clinical isolates irrespective of source produced substantial levels of enzymes assayed, produced melanin and exhibited swimming and swarming motility pattern. These data suggests clinical isolates of S. maltophilia is a reservoir for pathogenic potential enzymes.
Stenotrophomonas maltophilia plays a major role as an opportunistic pathogen in immunocompromised individuals. Despite not yet being recorded as a highly virulent bacterium, S. maltophilia is often isolated from serious human infection.Citation1 High rates of isolation in immunocompromised patients, increasing multidrug-resistant strains, and lack of controlled clinical treatment trials makes this bacterium a cause of serious concern.Citation2-Citation5S. maltophilia is frequently isolated from clinical specimens and is implicated in catheter-related bacteremia and septicemia, urinary and respiratory tract infections, and endocarditis.Citation6-Citation8 In most cases, S. maltophilia cause infection in patients with the assistance of invasive medical devices than in healthy host.Citation9
Despite its clinical relevance, very little is known about the pathogenic mechanism of infections. Secretion of enzymes, namely proteinases, lecithinases, gelatinase, lipases, and DNase, biofilm formation, hemolysis, and motility are few properties that pathogenic microbes possess in order to establish an infection. These enzymes are considered virulent as they damage the host tissues making the host permissive for infection.Citation10,Citation11
Although studies have implicated the ability of S. maltophilia to attach to mammalian cells, form biofilms and produce large number of extracellular enzymes,Citation12-Citation14 the presence of these factors in clinical isolates and its correlation with isolates recovered from different anatomical sites are not clear. Therefore, the main aims of the present study were to screen for the production of extracellular enzymes (protease, lipase, lecithinase, gelatinase, deoxyribonuclease [DNase], hyaluronidase, hemolysin) by S. maltophilia isolates, to compare the enzyme profiles of invasive (e.g., blood and cerebrospinal fluid) and non-invasive (e.g., sputum, tracheal aspirate, wound infection, and urine) isolates and also in device and non-device related isolates.
A total of 108 S. maltophilia clinical isolates collected from patients admitted for various underlying diseases in tertiary care hospitals in the central region of Malaysia were investigated. Isolates were confirmed as S. maltophilia by lavender green colonies on blood agar, motility test, and standard biochemical assays such as DNase, catalase and oxidase activities. The isolates were genotypically confirmed by species specific polymerase chain reaction (PCR) that targeted the 23s RNA. The isolates were further reconfirmed using the VITEK® Mass Spectrometry System and an array of 64 biochemical assays. S. maltophilia ATCC 13637 (clinical strain) and S. maltophilia LMG 959 (environmental strain) from Belgian Coordinated Collections of Microorganisms (BCCM) was used as control. These study isolates were further classified as invasive or non-invasive based on the anatomical site of isolation. Invasive isolates included in the study were those that were isolated from sites such as peripheral blood and cerebrospinal fluid. Non-invasive isolates were those that were isolated from sites such as tracheal aspirate, sputum wound infections, swabs, and pus.
All isolates were screened for the secretion of a panel of extracellular enzymes such as DNase, gelatinase, hemolysin, heparinase, hyaluronidase, lecithinase, lipase, and proteinase; pigment production, which includes melanin, fluorescein, and pyocyanin; and ability to form biofilm and motility patterns. The enzyme activity was evaluated by spot (2-mm) inoculations. All substrate plate assays were incubated for 48 h at 37 °C unless otherwise specified.
For DNase agar test, 0.01% toluidine blue was used to determine DNase production after 72 h of growth at 37 °C. DNase activity was indicated by the formation of a large pink halo around an inoculum spot.Citation15 Modified DNase tube test was also employed to evaluate the DNase production as described earlier. Clearing of the genomic DNA band was considered to be a confirmatory test.Citation16 For gelatinase test, organisms were inoculated on 0.4% gelatin agar. The plates were incubated at 37 °C for 24 h followed by which the plates were flooded with mercuric chloride solution. Appearance of opaque zone around the inoculum showed positive results.Citation17,Citation18 Hemolysis was determined on trypticase soy agar containing 5% sheep blood.Citation19 Heparinase production was tested by diluting heparin in distilled water to a final concentration of 5 U/ml followed by filter sterilization (0.45 pM) before dispensing 20 μl into 96-well micro titration plate; each well contained 30 μl of the test bacteria, incubated overnight at 37 °C. Twenty microliters of aqueous toluidine blue 0.01% was added to each well. Blue color indicated positive result, while pink indicated negative.Citation20 Hyaluronidase production was evaluated by incorporation of aqueous solutions of hyaluronic acid into Muller Hinton agar supplemented with bovine serum albumin (final concentration, 1%). After being inoculated and incubated for 48 h, each plate was flooded with 2 N acetic acid, which was removed after 10 min. The appearance of a clear zone around the inoculum indicated presence of hyaluronidase.Citation21 For lecithinase, 10 ml of the 50% egg yolk was added to 90 ml of sterilized tryptic soya agar. A white precipitate around or beneath an inoculum spot indicated lecithinase formation.Citation10,Citation22 Lipase activity was detected by the appearance of a turbid halo around the inocula on trypticase soy agar plates supplemented with 1% Tween 80.Citation23 Proteinase activity was evaluated using casein hydrolysis and was tested on Mueller–Hinton agar containing 3% (w/v) skimmed milk . The presence of a transparent zone around the inoculum spot indicated a positive test.Citation10,Citation24
For melanin production, bacteria were grown on l-tyrosine containing agar plates to observe the brown pigment produced.Citation25 Brown color pigmentation is considered positive. Pyocyanin was tested by using Bacto Pseudomonas Agar P. The pyocyanin production is promoted by glycerol which is the source of energy while magnesium chloride and potassium sulfate enhances the intensity of pyocyanin produced.Citation26,Citation27 Positive results were indicated by a bluish-green pigment that diffuses into the agar under UV light (366 nm).Citation10 For fluorescein, Bacto Pseudomonas Agar F is utilized. The presence of dipotassium phosphate increases the phosphorous content and magnesium sulfate provides essential ions whereby enhances the fluorescein production.Citation27 Under UV light (366 nm) a greenish yellow fluorescent pigment in the colonies and surrounding medium indicates fluorescein production.Citation10 For all pigments, the intensities were scored as 0, 1+, 2+, or 3+ manually in a semi-quantitative way. Intensity greater than 2+ was considered as high pigment-producing strain.Citation28
For biofilm assay the bacteria were grown overnight with agitation, the optical densities at 600 nm were standardized to 2, and the cultures were diluted 1:100 in Luria bertani (LB) broth. Aliquots (100 µl) were added to a 96-well plate, incubated for 24 h at 37 °C. Upon complete growth, the inoculums were discarded and washed three times with water. The plates were then incubated with crystal violet for 15 min, followed by 3 rinses with water (30 s each). Finally the biofilms were dissolved with 95% ethanol and transferred to fresh 96-well plate to measure the absorbance at 540 nm. Each sample was tested in triplicate.Citation29 The cutoff was defined as three standard deviations above the mean optical density of control (ODc).Citation30 Each isolate was classified as follows: weak biofilm producer OD ≤ 2 × ODc, moderate biofilm producer OD ≤ 4 × ODc and strong biofilm producer OD > 4 × ODc.Citation31
Motility patterns of S. maltophilia were determined by examining its ability to diffuse in soft agar plates. For swimming test, tryptone broth (10 g/l tryptone/5 g/l NaCl) that contained 0.3% (w/v) agarose was used. Swim plates were inoculated with bacteria from an overnight culture in LB agar (1.5%, w/v) plates at 37 °C with a sterile toothpick. The plates were then wrapped with saran wrap to prevent dehydration and incubated at 30 °C for 12–14 h. For swarming test, the media used consisted of 0.5% (w/v) bacto-agar with 8 g/l nutrient broth, to which 5 g/l glucose was added. Swarm plates were typically allowed to dry at room temperature overnight before being used. For twitching motility test, LB broth (10 g/l tryptone/5 g/l yeast extract/10 g/l NaCl) solidified with 1% (w/v) granulated agar was used. Twitch plates were briefly dried and isolates were stab inoculated with a sharp toothpick to the bottom of the petri dish from an overnight-grown LB agar (1.5%, w/v) plate. After incubation at 37 °C for 24 h, motility patterns were determined by measuring the zone of motility in the agar.Citation32 All experiments were conducted in triplicates. The data was analyzed using graph pad prism version 5.0. Values were considered significant when P < 0.05.
As summarized in enzymatic profile indicates that most of the isolates irrespective of being invasive or non-invasive secreted all of the major enzymes that are known to destroy cell components such as membranes and proteins. shows some of the assays that were tested for S. maltophilia isolates.
Table 1. Enzymatic profiles for clinical isolates from different anatomical sites as well as device-related and non-device-related isolates
Figure 1. Enzymatic profile of S. maltophilia isolates. (A) SM ATCC, (B) clinical isolate of S. maltophilia; (C) S. maltophilia LMG 959 (environmental isolate). When (A–C) were compared, for all tested properties, (C) (environmental strain) showed comparatively less activity.
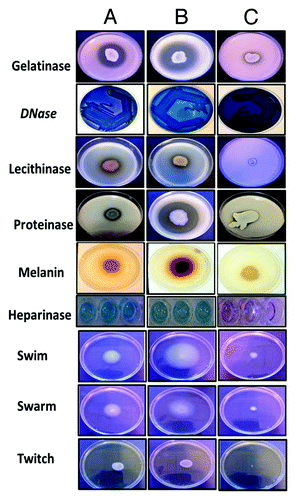
DNase, gelatinase, hemolysin, lipase, and proteinase activity were observed in all isolates studied. None of the urine isolates exhibited lecithinase, hyaluronidase, and heparinase activity. While isolates from most of the anatomical sites showed activity, 19.1% of tracheal aspirate and 56.5% of blood isolates, lecithinase secretion was observed to be absent. Among all enzymes assayed, heparinase showed variation based on the anatomical sites of isolates; 71.4% tracheal aspirate, 69.2% blood, 66.6% CSF, 80% sputum, and 69.2% wound infection were found to be positive. For pigment production, melanin was produced by majority of the invasive (91.1%) and non-invasive (95.2%). Pyocyanin and fluorescein were not observed by any of the isolates.
As depicted in , all isolates were biofilm producers with varied intensities. While majority of the isolates were low biofilm producers (80% invasive and 82.5% non-invasive), some showed a very strong biofilm formation (20% invasive and 17.5% non-invasive). With respect to motility, all isolates were found to be motile. Among the three modes of motility pattern tested, swimming and swarming were commonly seen, while twitching was not prominent.
Table 2. Production of melanin, biofilm, and motility profile of clinical isolates of S. maltophilia
When isolates were classified as device related and non-device related, as observed for anatomical sites, most of the enzymes were secreted by all isolates. Heparinase was found to be produced by slightly more number of device-related isolates (73.2%) than non-device-related ones (62.1%). In contrast, lecithinase was observed more among non-device-related isolates than device-related ones. However, both enzymes did not show any significant (P > 0.05) association with source of isolation. As observed in , significant association was also not observed in melanin and biofilm formation.
The striking increase in S. maltophilia infections with broad spectrum of clinical syndromes and with greater risk of death especially among immunocompromised and hospitalized patients has prompted an intense search for the underlying virulence determinants. To date, virulence factors of S. maltophilia is very minimally defined. It is very certain that S. maltophilia which is basically an environmental microbe would express pool of virulence determinants to establish itself as a successful clinical pathogen. Here, we performed a comprehensive screening of pathogenic determinants and extracellular enzymes secretion in clinical isolates of S. maltophilia. Our results clearly indicated that unlike environmental strain (LMG 959) clinical isolates of S. maltophilia is capable of producing a variety of extracellular enzymes and most of these enzymes are of known pathogenic potential.Citation33,Citation34
Proteinases plays an important role in invasiveness, host tissue damage, and evading host-defense.Citation35 Extracellular lipases help bacteria to thrive in a carbohydrate restricted environment where lipids are the sole carbon sourceCitation23,Citation36 and enable in adhering to host tissue.Citation37 Lecithinase enzyme modulates the host immune systemCitation38 and play roles in cell-to-cell spread as seen in Listeria monocytogenes pathogenicity.Citation39 Hyaluronidase facilitates tissue invasion.Citation40 Heparinase plays import role in the digestion of extracellular matrix of the host tissues (mainly related to the respiratory organs) which enhances the invasiveness of the pathogenCitation41 DNase evade the innate immune response in the host by degrading neutrophil extracellular traps (NETs) and also has the capability to kill bacteria by interfering with DNA synthesis.Citation42,Citation43
Although S. maltophilia is frequently associated with respiratory infections, like most other nosocomial pathogens, it is also isolated from different body sites or clinical specimens such as blood, urine, wound swabs, sputum, CSF, etc. The growth of S. maltophilia from non-sterile sites is sometimes difficult to interpret and is not used as a proof of infection. Hence isolation from sterile sites (e.g., blood, CSF), usually represents true infection. In this study we compared invasive (e.g., blood, CSF), and non-invasive (urine, tracheal aspirate, wound swab, sputum) isolates for the production of pathogenic determinants. Among all enzymes tested lecithinase, hyaluronidase, and heparinase were not detected in urine sample. Heparinase activity was found more among sputum and tracheal aspirate. Heparinase bind with heparin sulfate (substrate for heparinase) which is a component of the proteoglycans present in bronchial airways in the extracellular matrix and transmit pathologic processes leading to tracheobronchial infection.Citation44
Production of pigments like melanin protects bacteria from host defense mechanism and against environmental insults.Citation45 Although S. maltophilia is an inevitable pathogen among the immunocompromised patients, melanin production and the ability to form biofilms may enhance its protection from host defense mechanism. Melanin production and biofilm forming property has also shown to have association with antibiotic resistance in S. maltophilia.Citation46
S. maltophilia infection is usually mediated by the device or lines attached to the patients, for example breathing tubes such as endotracheal or tracheostomy tubes, ventilators and indwelling urinary catheters. Hence we compared isolates from device related and non-device related infectious to observe any differences in the virulence determinant profiles. No significant difference in any of the virulence determinant tested indicated, all clinical isolates could be reservoir for pathogenic potential determinants.
In conclusion, from the present study, it is clear that S. maltophilia irrespective of whether invasive or non-invasive, device-related or non-device-related is capable of producing virulence potential extracellular enzymes, which might play major roles in pathogenicity.
| Abbreviations: | ||
| NETs | = | neutrophil extracellular traps |
| DNase | = | deoxyribonuclease |
| CSF | = | cerebrospinal fluid |
| UV | = | ultraviolet |
| PCR | = | polymerase chain reaction |
| LB | = | Luria bertanii |
| OD | = | optical density |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Looney WJ, Narita M, Mühlemann K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis 2009; 9:312 - 23; http://dx.doi.org/10.1016/S1473-3099(09)70083-0; PMID: 19393961
- Safdar A, Rolston KV. Stenotrophomonas maltophilia: changing spectrum of a serious bacterial pathogen in patients with cancer. Clin Infect Dis 2007; 45:1602 - 9; http://dx.doi.org/10.1086/522998; PMID: 18190323
- Rolston KV, Kontoyiannis DP, Yadegarynia D, Raad II. Nonfermentative gram-negative bacilli in cancer patients: increasing frequency of infection and antimicrobial susceptibility of clinical isolates to fluoroquinolones. Diagn Microbiol Infect Dis 2005; 51:215 - 8; http://dx.doi.org/10.1016/j.diagmicrobio.2004.11.002; PMID: 15766609
- Andrews JM, BSAC Working Party on Susceptibility Testing. BSAC standardized disc susceptibility testing method (version 7). J Antimicrob Chemother 2008; 62:256 - 78; http://dx.doi.org/10.1093/jac/dkn194; PMID: 18474513
- Nicodemo AC, Paez JI. Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur J Clin Microbiol Infect Dis 2007; 26:229 - 37; http://dx.doi.org/10.1007/s10096-007-0279-3; PMID: 17334747
- Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev 1998; 11:57 - 80; PMID: 9457429
- Looney WJ. Role of Stenotrophomonas maltophilia in hospital-acquired infection. Br J Biomed Sci 2005; 62:145 - 54, quiz 1, 154; PMID: 16196464
- Senol E. Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J Hosp Infect 2004; 57:1 - 7; http://dx.doi.org/10.1016/j.jhin.2004.01.033; PMID: 15142709
- Araoka H, Baba M, Yoneyama A. Risk factors for mortality among patients with Stenotrophomonas maltophilia bacteremia in Tokyo, Japan, 1996-2009. Eur J Clin Microbiol Infect Dis 2010; 29:605 - 8; http://dx.doi.org/10.1007/s10096-010-0882-6; PMID: 20177726
- Edberg S, Gallo P, Kontnick C. Analysis of the virulence characteristics of bacteria isolated from bottled, water cooler, and tap water. Microb Ecol Health Dis 1996; 9:67 - 77; http://dx.doi.org/10.3109/08910609609166445
- Pavlov D, de Wet CM, Grabow WO, Ehlers MM. Potentially pathogenic features of heterotrophic plate count bacteria isolated from treated and untreated drinking water. Int J Food Microbiol 2004; 92:275 - 87; http://dx.doi.org/10.1016/j.ijfoodmicro.2003.08.018; PMID: 15145586
- De Vidipó LA, De Marques EA, Puchelle E, Plotkowski MC. Stenotrophomonas maltophilia interaction with human epithelial respiratory cells in vitro. Microbiol Immunol 2001; 45:563 - 9; http://dx.doi.org/10.1111/j.1348-0421.2001.tb01287.x; PMID: 11592630
- Pompilio A, Crocetta V, Confalone P, Nicoletti M, Petrucca A, Guarnieri S, Fiscarelli E, Savini V, Piccolomini R, Di Bonaventura G. Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol 2010; 10:102; http://dx.doi.org/10.1186/1471-2180-10-102; PMID: 20374629
- Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 2008; 9:R74; http://dx.doi.org/10.1186/gb-2008-9-4-r74; PMID: 18419807
- Janda JM, Bottone EJ. Pseudomonas aeruginosa enzyme profiling: predictor of potential invasiveness and use as an epidemiological tool. J Clin Microbiol 1981; 14:55 - 60; PMID: 6790569
- Neela V, Thomas R, Rankouhi SZ, Karunanidhi A, Shueh CS, Hamat RA, van Belkum A. Modified DNase tube test to detect DNase activity in Stenotrophomonas maltophilia. J Med Microbiol 2012; 61:1792 - 4; http://dx.doi.org/10.1099/jmm.0.049403-0; PMID: 22956752
- McDADE JJ, Weaver RH. Rapid methods for the detection of gelatin hydrolysis. J Bacteriol 1959; 77:60 - 4; PMID: 13620650
- Frazier WC. A Method for the Detection of Changes in Gelatin Due to Bacteria Two Plates. J Infect Dis 1926; 39:302 - 9; http://dx.doi.org/10.1093/infdis/39.4.302
- Travassos LH, Pinheiro MN, Coelho FS, Sampaio JL, Merquior VL, Marques EA. Phenotypic properties, drug susceptibility and genetic relatedness of Stenotrophomonas maltophilia clinical strains from seven hospitals in Rio de Janeiro, Brazil. J Appl Microbiol 2004; 96:1143 - 50; http://dx.doi.org/10.1111/j.1365-2672.2004.02248.x; PMID: 15078532
- Riley T. A simple method for detecting heparinase‐producing bacteria. Lett Appl Microbiol 1987; 4:21 - 3; http://dx.doi.org/10.1111/j.1472-765X.1987.tb01573.x
- Smith RF, Willett NP. Rapid plate method for screening hyaluronidase and chondroitin sulfatase-producing microorganisms. Appl Microbiol 1968; 16:1434 - 6; PMID: 4877667
- Nord CE, Sjöberg L, Wadström T, Wretlind B. Characterization of three Aeromonas and nine pseudomonas species by extracellular enzymes and haemolysins. Med Microbiol Immunol 1975; 161:79 - 87; http://dx.doi.org/10.1007/BF02121748; PMID: 1170482
- Rollof J, Hedström SA, Nilsson-Ehle P. Lipolytic activity of Staphylococcus aureus strains from disseminated and localized infections. Acta Pathol Microbiol Immunol Scand B 1987; 95:109 - 13; PMID: 3591310
- Burke V, Robinson JO, Richardson CJ, Bundell CS. Longitudinal studies of virulence factors of Pseudomonas aeruginosa in cystic fibrosis. Pathology 1991; 23:145 - 8; http://dx.doi.org/10.3109/00313029109060814; PMID: 1745565
- Wang G, Aazaz A, Peng Z, Shen P. Cloning and overexpression of a tyrosinase gene mel from Pseudomonas maltophila. FEMS Microbiol Lett 2000; 185:23 - 7; http://dx.doi.org/10.1111/j.1574-6968.2000.tb09035.x; PMID: 10731602
- King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 1954; 44:301 - 7; PMID: 13184240
- MacFaddin J. Media for the isolation-cultivation-identification-maintenance of medical bacteria, vol. 1 Williams & Wilkins. Baltimore, MD 1985.
- Liaw S-J, Lee Y-L, Hsueh P-R. Multidrug resistance in clinical isolates of Stenotrophomonas maltophilia: roles of integrons, efflux pumps, phosphoglucomutase (SpgM), and melanin and biofilm formation. Int J Antimicrob Agents 2010; 35:126 - 30; http://dx.doi.org/10.1016/j.ijantimicag.2009.09.015; PMID: 19926255
- Passerini de Rossi B, Calenda M, Vay C, Franco M. Biofilm formation by Stenotrophomonas maltophilia isolates from device-associated nosocomial infections. Rev Argent Microbiol 2007; 39:204 - 12; PMID: 18390153
- Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol 1985; 22:996 - 1006; PMID: 3905855
- Stepanović S, Cirković I, Ranin L, Svabić-Vlahović M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol 2004; 38:428 - 32; http://dx.doi.org/10.1111/j.1472-765X.2004.01513.x; PMID: 15059216
- Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 2000; 97:4885 - 90; http://dx.doi.org/10.1073/pnas.060030097; PMID: 10758151
- Bottone EJ, Reitano M, Janda JM, Troy K, Cuttner J. Pseudomonas maltophilia exoenzyme activity as correlate in pathogenesis of ecthyma gangrenosum. J Clin Microbiol 1986; 24:995 - 7; PMID: 3537006
- O’Brien M, Davis GH. Enzymatic profile of Pseudomonas maltophilia. J Clin Microbiol 1982; 16:417 - 21; PMID: 6813350
- Travis J, Potempa J, Maeda H. Are bacterial proteinases pathogenic factors?. Trends Microbiol 1995; 3:405 - 7; http://dx.doi.org/10.1016/S0966-842X(00)88988-X; PMID: 8564361
- Stehr F, Kretschmar M, Kröger C, Hube B, Schäfer W. Microbial lipases as virulence factors. J Mol Catal, B Enzym 2003; 22:347 - 55; http://dx.doi.org/10.1016/S1381-1177(03)00049-3
- Miskin JE, Farrell AM, Cunliffe WJ, Holland KT. Propionibacterium acnes, a resident of lipid-rich human skin, produces a 33 kDa extracellular lipase encoded by gehA. Microbiology 1997; 143:1745 - 55; http://dx.doi.org/10.1099/00221287-143-5-1745; PMID: 9168624
- Cheng Q, Yu MC, Reeves AR, Salyers AA. Identification and characterization of a Bacteroides gene, csuF, which encodes an outer membrane protein that is essential for growth on chondroitin sulfate. J Bacteriol 1995; 177:3721 - 7; PMID: 7601836
- Vazquez-Boland JA, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun 1992; 60:219 - 30; PMID: 1309513
- Duran-Reynals F. Studies on a Certain Spreading Factor Existing in Bacteria and Its Significance for Bacterial Invasiveness. J Exp Med 1933; 58:161 - 81; http://dx.doi.org/10.1084/jem.58.2.161; PMID: 19870187
- Lodise TP, McKinnon PS. Clinical and economic impact of methicillin resistance in patients with Staphylococcus aureus bacteremia. Diagn Microbiol Infect Dis 2005; 52:113 - 22; http://dx.doi.org/10.1016/j.diagmicrobio.2005.02.007; PMID: 15964499
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science 2004; 303:1532 - 5; http://dx.doi.org/10.1126/science.1092385; PMID: 15001782
- Fox JB, Holtman DF. Effect of anaerobiosis on staphylococcal nuclease production. J Bacteriol 1968; 95:1548 - 50; PMID: 5650065
- Rahmoune H, Lamblin G, Lafitte JJ, Galabert C, Filliat M, Roussel P. Chondroitin sulfate in sputum from patients with cystic fibrosis and chronic bronchitis. Am J Respir Cell Mol Biol 1991; 5:315 - 20; http://dx.doi.org/10.1165/ajrcmb/5.4.315; PMID: 1910815
- Nosanchuk JD, Casadevall A. The contribution of melanin to microbial pathogenesis. Cell Microbiol 2003; 5:203 - 23; http://dx.doi.org/10.1046/j.1462-5814.2003.00268.x; PMID: 12675679
- Liaw SJ, Lee YL, Hsueh PR. Multidrug resistance in clinical isolates of Stenotrophomonas maltophilia: roles of integrons, efflux pumps, phosphoglucomutase (SpgM), and melanin and biofilm formation. Int J Antimicrob Agents 2010; 35:126 - 30; http://dx.doi.org/10.1016/j.ijantimicag.2009.09.015; PMID: 19926255
