Abstract
Some of the mechanisms underlying the invasion and intracellular survival of B. melitensis are still unknown, including the role of a subfamily of NUDIX enzymes, which have been described in other bacterial species as invasins and are present in Brucella spp. We have generated a mutation in the coding gene of one of these proteins, the invA gene (BMEI0215) of B. melitensis strain 133, to understand its role in virulence. HeLa cell invasion results showed that mutant strain survival was decreased 5-fold compared with that of the parental strain at 2 h pi (P < 0.001). In a goat macrophage infection assay, mutant strain replication was 8-fold less than in the parental strain at 24 h pi (P < 0.001); yet, at 48 h pi, no significant differences in intracellular replication were observed. Additionally, colocalization of the invA mutant with calregulin was significantly lower at 24 h pi compared with that of the parental strain. Furthermore, the mutant strain exhibited a low level of colocalization with cathepsin D, which was similar to the parental strain colocalization at 24 h pi. In vivo infection results demonstrated that spleen colonization was significantly lower with the mutant than with the parental strain. The immune response, measured in terms of antibody switching and IFN-γ transcription, was similar for Rev1 and infection with the mutant, although it was lower than the immune response elicited by the parental strain. Consequently, these results indicate that the invA gene is important during invasion but not for intracellular replication. Additionally, mutation of the invA gene results in in vivo attenuation.
Introduction
Brucellosis is a worldwide zoonotic disease that is mainly distributed in undeveloped countries. In animals, infection with the causative bacterium leads to reproductive problems, abortion and lesions in a variety of tissues, resulting in chronic illness.Citation1 Brucellosis caused by B. melitensis is controlled in animals by the Rev1 vaccine.Citation2 The Brucella genus lacks classical virulence mechanisms, and its metabolism is closely linked to its intracellular lifestyle.Citation3 B. melitensis combats the harsh intracellular environment by transcribing several genetic factors in order to inhibit intracellular destruction and promote bacterial multiplication.Citation4-Citation6 It has been demonstrated that Brucella is capable of modulating the transcription of metabolic genes (for processes such as carbon, nitrogen and lipid metabolism) depending on the environmental conditions.Citation7 A crucial step for intracellular survival is the invasion process, during which Brucella spp. must deal with oxidative and nutrient stress.Citation6,Citation8 A metabolic shift known as the stringent response occurs under nutrient starvation during the invasion process and is characterized by inhibition of RNA and ribosomal protein synthesis.Citation9 These conditions raise the intracellular concentration of recognized molecules known as alarmones (oligophosphate nucleotides). This increase in alarmone concentration is interpreted by the cell as a signal alerting the cell to prepare for a stress adaptation with a transcriptional change.Citation10-Citation12 Although initially beneficial, the accumulation of alarmones can also be detrimental to cells.Citation12,Citation13 To inhibit the toxic effects of alarmone accumulation, a special subfamily of NUDIX enzymes can hydrolyze these oligophosphates. The NUDIX enzymes are a diverse family of enzymes that act on (di)nucleoside oligophosphate molecules linked to other “X” molecules.Citation14 These enzymes have been described in R. prowazekii,Citation15 E. coli,Citation16 L. pneumophila,Citation17 S. Typhimurium,Citation18 and B. bacilliformis.Citation19 The exact link between the transcription of these enzymes and their role in virulence is not fully understood; however, they may act to reduce stress-induced alarmone levels, promoting invasion and intracellular survival.Citation15,Citation19
In the B. melitensis genome, the BMEI0215 (invA) gene has phosphate nucleoside hydrolase characteristics. In an amino acid analysis, the Brucella melitensis invA gene had 52% similarity to NUDIX enzymes described in other pathogenic bacteria (). This gene exhibited the closest similarity to the invA gene of Rickettsia prowazekii (GenBank access no. RP236) and the ialAB locus of Bartonella bacilliformis (nudH-ialB locus), with an average of 76%.
Figure 1. Alignment of the Brucella melitensis invA amino acid sequence (BMEI0215) and homologous invasive bacterial proteins in a BLAST search. The NUDIX signature sequence is shown in the black square. Identical amino acids and similar amino acids are indicated in dark shaded and light shaded boxes, respectively.
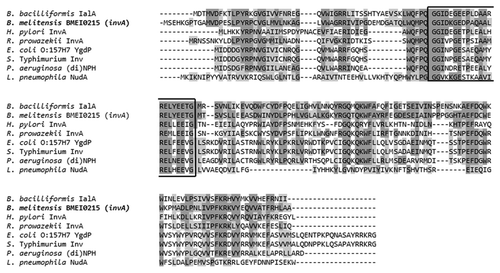
Given the importance of stress adaptation, the role of the invA gene could be crucial in the virulence of B. melitensis, as has been demonstrated for other pathogenic bacteria. However, to our knowledge, no studies of the BMEI0215 (invA) gene have been performed in B. melitensis.
In the present work, we have shown that expression of the invA gene could be important for intracellular invasion and virulence in a murine model of infection.
Results
HeLa cell invasion
Invasion was analyzed by infecting epithelial-like HeLa cells with different B. melitensis strains at an MOI of 500 and sampling the cultures at different times pi (). At the onset of infection (incubation time zero), all the strains demonstrated an invasion average of 4.72 × 106 CFU/mL, except for B. melitensis 16M, for which there were 2.2-fold more bacteria present than compared with the other strains (P < 0.05). At 30 min pi, B. melitensis 16M (Bm 16M, reference strain) exhibited 1.6-fold greater survival than B. melitensis 133 (P < 0.05, Bm 133, parental) and 4-fold greater survival than B. melitensis 133 invA-km (Bm A, mutant strain), B. melitensis 133 invA-kmC (Bm AC, complemented strain), and B. melitensis Rev1 (Bm Rev1, vaccine strain) (P < 0.001). At the same time pi, Bm 133 survival was 2.5-fold greater than that of Bm A, Bm AC, and Bm Rev1 (P < 0.05). Later, at 60 min pi, no significant differences were observed for the survival of the strains. An increase in bacterial concentration for Bm 16M, Bm 133, and Bm AC was evident at 120 min pi; in contrast, Bm Rev1 and Bm A retained the same levels of intracellular bacteria compared with 60 min pi. This bacterial increase might be attributable to the Brucella duplication time. It is known that virulent strains of Brucella in log-phase have a duplication time of 2 to 2.5 h in culture media.Citation20,Citation21 It is probable that virulent strains could be duplicating during the invasion period. The survival of the Bm A and Rev1 strains at 120 min pi was similar (P > 0.05) and was 5-fold less than the bacterial concentrations of the parental, reference, and complemented strains (P < 0.001, ).
Figure 2. Intracellular invasion and survival in HeLa cells. The results are shown as colony forming units (CFU) at different times pi. Significant differences are shown as black stars (P < 0.05) or gray stars (P < 0.001). Lines and symbols represent: solid line and closed circle, B. melitensis 133; dashed line and open triangle, B. melitensis 133 invA-km; dashed line and open inverted triangle, B. melitensis 133 invA-kmC; dashed line and closed diamond, B. melitensis Rev1; solid line and open circle, B. melitensis 16M. The results are the average of three experiments ± standard deviation.
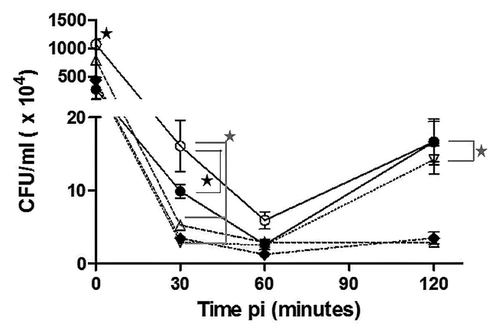
The decreased bacterial concentration observed at 120 min pi for the mutant and vaccine strains could be related to their attenuated phenotypes. Although the complemented mutant strain was similar to the parental strain at the final time point, there was no effect before 120 min pi, suggesting partial complementation of the strain ().
Survival and intracellular multiplication in goat macrophages
The intracellular survival of different Brucella strains was measured at different time points in goat macrophages infected at an MOI of 100. At 0 and 4 h pi, no significant differences were evident between the strains in terms of invasion and intracellular survival, with the exception of the Bm Rev1 strain, which exhibited 2.5-fold lower survival than the others at 4 h pi (P < 0.05, ). At 24 h pi, the parental and reference strains survived and replicated uniformly, at higher concentrations, i.e., 11-fold greater than the rest of the strains tested (P < 0.001). At 48 h pi, the parental, reference, and mutant strains demonstrated 16-fold greater replication than the Rev1 and complemented strains (P < 0.001). During the phagocytosis (0 h) and adaptation (4 h) periods, there were no significant differences between the mutant strain and the parental strain (P > 0.05). At 24 h pi, the mutant strain demonstrated 8-fold less replication than the parental strain, which was the most pronounced difference in terms of the intracellular concentration of both strains (P < 0.001). In contrast, no significant differences (P > 0.05) were observed between the parental strain and the mutant strain at 48 h pi, indicating that the invA mutation results in delayed multiplication. As seen in , complementation did not restore the survival and replication ability of the mutant strain at 24 h and 48 h pi. In fact, the Bm AC strain demonstrated 4.2-fold lower levels of survival than the Bm A strain at 24 h pi and 6.2-fold lower levels at 48 h pi, indicating less survival and intracellular replication of the complemented mutant strain than of the mutant strain (P < 0.05).
Figure 3. Phagocytosis and intracellular survival in caprine macrophages. The results are shown as colony forming units (CFU) at different times pi. Significant differences are shown as black stars (P < 0.05) or gray stars (P < 0.001). Lines and symbols represent: solid line and closed circle, B. melitensis 133; dashed line and open triangle, B. melitensis 133 invA-km; dashed line and open inverted triangle, B. melitensis 133 invA-kmC; dashed line and closed diamond, B. melitensis Rev1; solid line and open circle, B. melitensis 16M. The results are the average of 3 experiments ± standard deviation.
Intracellular traffic in murine macrophages
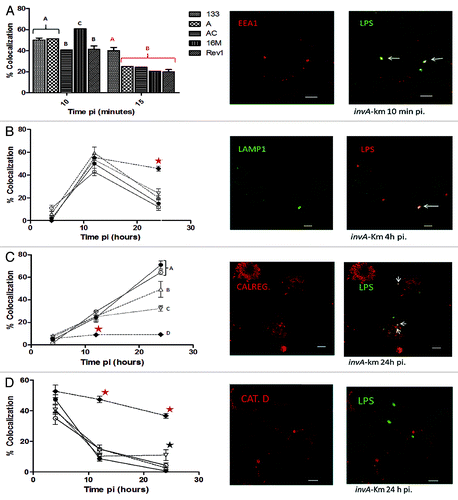
Infection of the murine macrophage J774A.1 cell line at a bacterial MOI of 100 was performed to study intracellular traffic via confocal microscopy using selected protein markers (). Confocal microscopy analysis revealed a similar entry pattern for all strains tested, with some differences at 10 min pi (), indicating that the reference strain had the highest percentage of EEA1 (early endosomal antigen 1) colocalization (60.6% ± 2%, P < 0.05). EEA1 is a protein that characterizes early endosomes and is involved in the phagosome to early-endosome transition.Citation22 Brucella-containing vacuoles (BCVs) of the parental strain retained the EEA1 protein (40% ± 2%, ) at 15 min pi, resulting in the highest colocalization percentage compared with other bacterial strains (P < 0.001), suggesting sustained invasion by the parental strain. The association of BCVs with EEA1 is transient and occurs immediately after internalization.Citation23 During endosome maturation, BCVs next acquire the LAMP1 glycoprotein (lysosome associated-membrane protein-1), which is characteristic of late endosomes and lysosomes.Citation24 During the adaptation period (4 h pi) BCVs of the different tested strains exhibited a similar pattern of positive LAMP1 colocalization (). At 12 h pi, the strains demonstrated an average of 52% positive LAMP1 vacuoles, and at 24 h pi, 17.8% positive LAMP1 colocalization was observed, with the exception of the Bm Rev1 strain. The percentage of colocalization of the vaccine strain with LAMP1 at 24 h pi was 45.7% ± 2.08% (P < 0.05). Later during BCVs intracellular trafficking, replicative vacuoles are evident when phagosomes progressively acquire endoplasmic reticulum markers, such as calregulin, and lose late endosomal markers, such as LAMP1.Citation25 As expected, in the parental and reference strains, calregulin-positive BCVs progressively increased during infection () and were higher at 24 h pi. This infection time point (24 h pi) positively correlated with the onset of macrophage intracellular replication (). The colocalization of calregulin in the Bm 133 and Bm 16M strains was similar (71% ± 3.6% and 64.3% ± 2%, respectively) at 24 h pi. These percentages were significantly higher than the calregulin-positive BCVs of the mutant strain (49.3% ± 7%, P < 0.05, ). As demonstrated by the macrophage infection results (), complementation of the mutant strain was only partial, as colocalization with calregulin at 24 h pi in the complemented mutant strain was significantly lower than in the parental strain (32.2% ± 2.5% vs. 71% ± 3.6%, respectively; P < 0.001). To measure intracellular destruction, a cathepsin D antibody was utilized (lysosomal protein). Replicative Brucella never colocalizes with cathepsin D, in contrast with attenuated bacteria.Citation24 Cathepsin D colocalization progressively decreased over the course of infection for all strains (), with the exception of Bm Rev1 at 12 h pi in comparison with the rest of the strains tested (47.3% ± 2.08% vs. 12.2% ± 0.6%, respectively; P < 0.001) and at 24 h pi (36.7% ± 1.5% vs. 4.7% ± 1.3%, respectively; P < 0.001). In agreement with the results for LAMP1-negative and calregulin-positive BCVs, for which the onset of intracellular replication was observed (, 24 h pi), it was expected that these strains would have a low percentage of cathepsin D-positive BCVs at 24 h pi. With the exception of the complemented mutant strain (11% ± 3.5%, P < 0.05), the parental, reference and mutant strains yielded similar cathepsin D colocalization results (an average of 7.5% ± 1.6%). These results indicated that the intracellular destruction of the strains at 24 h pi was not as great as that of the Bm Rev1 strain ().
Figure 4. Percentage of colocalization ± standard deviation of the B. melitensis strains with different endosomal antibodies (left) and representative confocal micrographs of the B. melitensis 133 invA-km mutant strain (right) at different times pi in J774A.1 macrophages. (A, C, and D): LPS colocalization (Alexa 488) with EEA1, calregulin, and cathepsin D (Alexa 594), respectively. (B) LPS colocalization (Alexa 594) with LAMP1 (Alexa 488). Bottom bar in micrographs corresponds to 5 μm; white arrows show colocalization. Significant differences are shown in letters and stars (black star represents a difference of P < 0.05, and red star represents a difference of P < 0.001). Lines and symbols represent: solid line and closed circle, B. melitensis 133; dashed line and open triangle, B. melitensis 133 invA-km; dashed line and open inverted triangle, B. melitensis 133 invA-kmC; dashed line and closed diamond, B. melitensis Rev1; solid line and open circle, B. melitensis 16M.
Residual virulence in the murine model
To analyze residual virulence, different groups of mice were inoculated with B. melitensis strains (1 × 104 CFU) intraperitoneally. Spleen colonization was evident at the beginning of murine infection (). At 7 d pi, mice inoculated with the parental strain achieved the highest levels of spleen colonization, with a tissue concentration of 498.6 ± 44.5 CFU/mg, which was 1.7-fold greater than that of the other strains (P < 0.001). The highest levels of spleen colonization were achieved at 15 d pi: 648.6 ± 153.3 CFU/mg for the parental group vs. the group infected with the mutant strain (480.3 ± 155.5 CFU/mg, P < 0.05), for which spleen colonization was 0.74-fold less than that of the parental group. Conversely, the Bm Rev1 and Bm AC groups demonstrated 6.2-fold less spleen colonization than the Bm A group (P < 0.001) and 8.5-fold less spleen colonization than the Bm 133 group (P < 0.001). Infection control began at 30 d pi; this was evident in the low bacterial recovery from mouse spleens infected with the Rev1, mutant and complemented strains (55.5 ± 19.5 CFU/mg, P > 0.05). At 30 d pi, the bacterial concentrations in spleens from the parental group were 6-fold greater than for the mutant, complemented mutant and Rev1-inoculated groups (P < 0.001); later, at 60 d pi, the recovery of the parental strain was 9.51-fold greater than for the mutant and complemented mutant groups (P < 0.001). Finally, at 90 d pi, the bacterial concentration for the parental group was 5.64-fold greater than that of the mutant group (P < 0.001). There was no bacterial recovery from spleens of either from mice inoculated with the Rev1 strain starting at 30 d pi through to the end of the experiment or mice in the complemented mutant group at 90 d pi.
Figure 5. Recovery of bacteria from spleens of infected mouse groups. The results are shown as colony forming units (CFU) per milligram of spleen at different times pi. Lines and symbols represent: solid line and closed circle,B. melitensis 133; dashed line and open triangle, B. melitensis 133 invA-km; dashed line and open inverted triangle, B. melitensis 133 invA-kmC; dashed line and closed diamond, B. melitensis Rev1. The results are the average of three mice ± standard deviation at different times pi.
The spleen weight of the different inoculated groups indicated active infection (). The most prominent splenomegaly was detected in all bacteria-inoculated groups during the first 15 d of infection. For the complemented mutant group, there was a constant spleen weight from day 30 pi to the end of the experiment (an average of 195.86 ± 41.36 mg), and constant splenomegaly was observed at 60 d and 90 d pi for the mutant group (220.97 ± 22.05 mg and 214.03 ± 40.93 mg, respectively). As seen in , the parental group had the highest spleen weights throughout the experiment until 30 d pi. At 30 d pi, the spleen weights for the parental and mutant groups were similar (446.1 ± 36.57 mg and 396.53 ± 54.5 mg, respectively; P > 0.05), and were 2.16-fold greater than the spleen weights of the Rev1 and complemented mutant groups (P < 0.001). Interestingly, after 60 d of infection, the spleen weights of the mutant and complemented groups had similar values (P > 0.05), likely indicating similar patterns of inflammation during late in vivo infection. Meanwhile, splenomegaly caused by the parental strain was 1.96-fold greater than that elicited by the mutant and complemented strains and 2.74-fold greater than that elicited by the Rev1 strain. Finally, at 90 d pi, splenomegaly caused by the parental strain was 1.96-fold greater than caused by the mutant, complemented and Rev1 strains. Taken together, the BALB/c mouse infection results demonstrated that, in vivo, the invA mutant strain was less virulent than the parental strain, although spleen colonization and splenomegaly indicated that the attenuation was not as evident as the Rev1 vaccine strain attenuation. Related to the in vitro results, the complemented mutant strain and the mutant strain exhibited similar virulence behaviors, indicating that complementation was partial.
Table 1. Spleen weights of infected mice on different days pi (d pi)
Humoral immune responses in the murine model
Sera from infected mice during the residual virulence assay were collected to determine isotype antibody switching at different time points throughout in vivo infection (). There was a general increase in antibody levels during the first 30 d pi. From 60 d to 90 d pi, there was a plateau for all antibodies tested, except for the IgM antibody titers. IgM antibodies were predominant over the course of infection (), but the IgG3 antibody titers were also high, indicating that they played a key role during infection (). During the first 7 d of infection for the group inoculated with the parental strain, the IgM antibody titers were predominant over the other IgG subclass antibodies (0.159 ± 0.082, ); however, negligible IgG2b () and IgG3 antibody titers () were detected during the same time period. In the group challenged with the parental strain, the IgM antibody titers increased at a constant rate throughout the experiment. However, in the mutant group, the IgM antibody titers were significantly higher than those elicited by the parental strain at 30 d pi (0.436 ± 0.03 vs. 0.270 ± 0.024, respectively; P < 0.001). IgG3 antibodies, as expected, were evident starting at 7 d pi in the parental group. These IgG antibodies predominanted over the other IgG subclasses in all groups. Differences in the IgG3 titers between the parental and mutant groups were perceptible at 30 d pi; the titers of the parental group were 1.3-fold greater than mutant titers (P < 0.001). Interestingly, the Rev1 group had similar IgG3 and IgG2b titers compared with the mutant group at 30 d pi, indicating similar humoral responses. IgG2b antibodies were more pronounced than IgG2a subclass antibodies, and were elicited earlier for the parental group (). At 30 (P < 0.001) and 90 (P < 0.05) days pi, the parental group exhibited significantly higher IgG2b antibody titers than the mutant group. As seen in , the parental group IgG2b titers always demonstrated values above the IgG2b titers of the mutant group, thus indicating that the humoral response was elicited to a considerably greater extent by the parental strain than by the mutant strain. However, the mutant IgG2a antibody titers demonstrated constant humoral stimulation beyond 15 d pi and were considerably higher at 30 d pi compared with those of the parental group (P < 0.001, ). In contrast to IgG2 antibodies, the IgG1 titers were markedly lower than the IgG2a and IgG2b titers in all inoculated groups. From 30 to 60 d pi, constant and similar IgG1 antibody levels were evident for the parental and mutant groups and at 90 d pi, a slight increase in parental IgG1 titers was significantly different from mutant IgG1 titers (P < 0.05, ).
Figure 6. Immune responses of infected mouse groups. (A–E) correspond to the humoral response. Results are expressed as the mean O.D. 490 ± standard deviation at different times pi. (A) IgG1 antibody results, (B) IgG2a, (C) IgG2b, (D) IgG3, and (E) IgM. (F) corresponds to IFN-γ mRNA transcription. IFN-γ expression was normalized to β-actin. Data are expressed as fold induction ± standard deviation compared with the control group (unstimulated). Bars represent the following: white bars, B. melitensis 133; black bars, B. melitensis 133 invA-km; light gray bars, B. melitensis Rev1; medium gray bars, B. melitensis 133 invA-kmC; dark gray bars, control group (unstimulated).
IgG2a and IgG2b antibodies are markers of the Th1 immune response; they are elicited to promote macrophage phagocytosis and IFN-γ production. In contrast, the IgG1 antibody is a typical marker of the Th2 immune response, which is characterized as the regulatory and humoral response.Citation26 In general, in the parental and mutant groups, the IgG2b or IgG2a antibody titers were dominant over the IgG1 antibody levels, indicating a Th1 immune response in in vivo infection. Conversely, in the complemented mutant group, antibody titers revealed a pattern that was similar to that elicited by the mutant strain, indicating in vivo plasmid instability as previously described.Citation27
Compared with the Rev1 group, the mutant group exhibited comparable antibody titers. There was no significant difference between IgM elicited by the Rev1 strain and that elicited by the mutant strain during the first 30 d pi. In relation to IgG antibodies, only the Rev1 IgG2b antibody titers were higher than those elicited by the mutant strain at 7 (0.084 ± 0.031 vs. 0.008 ± 0.005, P < 0.05) and 15 (0.145 ± 0.010 vs. 0.073 ± 0.02, P < 0.001) days pi.
IFN-γ expression in the murine model
mRNA samples were obtained from the blood of infected mice throughout the residual virulence assay to determine IFN-γ expression levels by real-time PCR (). As observed, IFN-γ transcription was upregulated during the first week pi for all inoculated groups. There was no difference in IFN-γ transcription between the parental, mutant and Rev1 inoculated mice, indicating that the Th1 polarization response was elicited similarly by these strains. After the first week pi IFN-γ transcription was inhibited, likely indicating that other cytokines were readily taking its place during in vivo infection.
Discussion
NUDIX hydrolases have been described in both prokaryotes, eukaryotes and even in viruses. They are implicated in several functions, including RNA processing, metabolic processes, genetic detoxification, and bacterial pathogenesis.Citation28 Their involvement in bacterial invasion was demonstrated with the E. coli YgdP protein,Citation16 IalA of B. bacilliformis,Citation19 and InvA of R. prowazekii.Citation15 The principal objective of the present work was to elucidate the role of a potential NUDIX (BMEI0215) gene in Brucella virulence. The epithelial-like cell invasion results demonstrated that survival of the B. melitensis invA-km strain was significantly compromised at 2 h pi, indicating the inability of this mutant strain to survive during the adaptation period of infection. Additionally, intracellular survival assay results with the caprine macrophage infection model revealed a considerable decrease in the intracellular concentration of the mutant strain at 24 h pi, which was 81% lower than the parental and reference strain concentrations. However, at 48 h pi, no significant concentration differences were observed between the mutant strain and the virulent strains, which indicate that the mutant strain maintains some degree of intracellular replication ability. In agreement with the caprine macrophage results, the colocalization results for cathepsin D revealed that there was no significant difference at 24 h pi. Conversely, the calregulin-positive BCV percentage for the mutant strain was less than the calregulin colocalization in the parental strain at the same time point (24 h pi), indicating lower overall intracellular multiplication of the mutant strain, which was not correlated with intracellular destruction (cathepsin D results). These findings show that the invA mutation restricts intracellular multiplication at 24 h pi. However, at 48 h pi, the intracellular concentration was not affected; multiplication of the mutant strain was similar to that of the Bm 133 and Bm 16M virulent strains. This observation could indicate a delayed intracellular multiplication of the mutant strain at 24 h pi but attainment of the same intracellular multiplication at 48 h as that of the other virulent strains, as has been observed for other Brucella virulence factors.Citation29,Citation30 These findings are in agreement with previous observations of mutated NUDIX enzymes in bacterial strains. Edelstein et al. showed that a L. pneumophila nudA mutant exhibited a significant decrease in guinea pig macrophage multiplication after 2 d pi; however, virulence restoration was evident on day 3 pi.Citation17 The invA gene likely acts as a factor that limits the accumulation of alarmones during the first stage of intracellular stress.Citation24 It is known that the accumulation of alarmones (such as diadenosine 5′,5′′′-P1,P4-tetraphosphate, Ap4A, or diadenosine 5′,5′′′-P1,P4 pentaphosphate, Ap5A) can be detrimental to intracellular bacteria; however, NUDIX hydrolases counteract the excess of alarmones to detoxify the intracellular niche.Citation13 It is probable that the invA gene of B. melitensis could restore homeostatic levels of these alarmones, indirectly promoting invasion. However, biochemical characterization of the InvA enzyme is crucial to determine the substrate specificity.
In contrast with the in vitro infection results, mutation of the invA gene of B. melitensis resulted in in vivo attenuation. At 15 d pi, mice infected with the mutant strain had high levels of bacterial spleen colonization compared with the levels in mice infected with the Rev1 vaccine and complemented strains; however, bacterial concentrations were not the same as they were for mice infected with the parental strain. At 15 d pi the mutant strain CFU concentration decreased to levels similar to those of the Rev1 vaccine strain, and those of the parental strain remained above the mutant and vaccine bacterial concentrations. Splenomegaly as a sign of inflammation can indicate bacterial colonization, as was evident in mice inoculated with the parental strain (). A constant decrease in bacterial concentration suggests control of the infection (). If we can link low bacterial concentrations with immune control, it is clear that immune control in the mutant-inoculated group was more effective than that observed in the parental strain group. The immune response in Brucella infection is principally mediated by a Th1 response.Citation26 The humoral response observed in the inoculated mice suggests this type of immune response, as IgG2a and IgG2b were predominant over IgG1 antibodies (). The principal differences in antibody titers between the parental and mutant group could be observed at 30 d pi; beyond this point, the differences were minimal. The parental strain elicited an earlier immune response than the mutant strain, as IgM and IgG3 antibodies were evident only in the parental inoculated group (). At 30 d pi, IgG2a was higher in the mutant inoculated group and IgG2b was lower in the mutant inoculated group, compared with the parental group. IgG1 antibody levels were evident during this time period, suggesting that a regulatory response was taking place after 30 d of infection. Similar antibody levels were detected starting at 30 d pi in the Rev1 group and the mutant group. In general, there was an analogous humoral response; however, differences between the parental and mutant inoculated groups could be indicators of specific immune responses. The representative cytokine of the Th1 response is IFN-γ, which exerts brucellicidal activity by activating macrophages.Citation26 As seen in , IFN-γ transcription was upregulated during the first week of infection for all inoculated groups. After 7 d pi, no IFN-γ transcription profile was evident, suggesting that B. melitensis control relies on other immune mechanisms. TNF-α and IL-2 are pro-inflammatory cytokines that participate in the Th1 immune response of BALB/c mice, in addition to IFN-γ.Citation31 It is possible that elevated TNF-α and IL-2 levels, as well as those of other cytokines, could mask IFN-γ transcription beyond 7 d pi. However, it is clear that there is no difference in IFN-γ transcription, suggesting that mutation of the invA gene elicits similar cellular immune responses as the parental strain at the beginning of infection. In vivo attenuation of the mutant strain could be related to intracellular destruction during the invasion process (as was observed in the HeLa assay) and, consequently, antigen presentation.
Survival, intracellular replication and in vivo virulence results for the complemented mutant strain (Bm 133 invA-kmC) indicated that complete restoration to wild-type levels was not possible. Genetic Brucella complementation in trans with the use of pBBR1MCS, a member of a moderate-copy number plasmid family, was shown to result in incomplete in vivo and in vitro restoration.Citation27,Citation32,Citation33 In the goat macrophage model, intracellular survival and replication of the complemented mutant strain were even lower than those of the mutant strain (). Similar effects were observed with Brucella ovis outer membrane protein (OMP) complemented mutants by Caro-Hernández et al. who observed that complemented strains were more susceptible to hydrogen peroxide, non-immune ram serum and selected drugs than mutant strains. In this study, the expression of OMPs in the complemented mutant strains was not identical as wild-type levels, suggesting that a certain level of OMP expression is necessary for membrane integrity.Citation33 Additionally, it has been reported that genetic complementation of the nudA gene of L. pneumophila and the invA gene of R. prowazekii in respective mutant strains does not restore their invasive capacity to wild-type levels; this is mainly attributed to the low production and effectiveness of the enzyme.Citation15,Citation17 Detection of InvA protein expression in the Bm 133 invA-kmC mutant will be important to determine whether its partial complementation is due to low protein expression. However, the mutation in the invA gene was created by interrupting the gene via the insertion of a kanamycin cassette from a promotorless pSU411 plasmid; a polar effect in adjacent genes was improbable. The use of the pBBRMCS1–4 plasmid for genetic complementation in this work, as well as the low production and effectiveness of plasmid transcription, likely resulted in incomplete phenotypic restoration.
We conclude that the invA gene is important for intracellular invasion and adaptation but is not necessary for intracellular replication. In vivo infection results suggest that the invA gene is required for full virulence of B. melitensis in the mouse model of infection. Indeed, mutation of the invA gene elicits a different humoral immune response but similar IFN-γ transcription levels during the first week of infection. The immune response is in agreement with bacterial recovery, as the peak in antibody titers at 30 d pi is related to the decrease in bacterial recovery. Functional gene redundancy in B. melitensis detoxification indicates that this bacterial species uses several mechanisms to avoid destruction inside the cell. These mechanisms include the Cu–Zn superoxide dismutase (SOD), AhpC (alkyl hydroxyperoxide reductase), Kat E (catalase), transcription of chaperones (dnaK, groEL, clpB), and several others.Citation6 Consequently, the invA gene may represent another mechanism that promotes invasion, such that mutation does not affect the intracellular replication of B. melitensis. Further research is required to evaluate the InvA enzymatic functionality and specificity.
Methods
Bacterial strains
The bacterial strains and plasmids used in the present work are shown in . The 133, Rev1, and 16M strains of B. melitensis were grown in Brucella broth and on Brucella agar (Becton Dickinson and Company, 211088 and 211086). The DH5α and S17.1 strains of E. coli were grown in Luria Bertani broth and on Luria Bertani agar (Becton, 241420 and 241320). When necessary, kanamycin (50 μg−1, Sigma Aldrich, 60615) and ampicillin (100 μg−1, Sigma Aldrich, A9393) were added. Genetic manipulations were performed according to standard procedures.Citation34
Table 2. Plasmids and bacterial strains used in this study
B. melitensis 133 invA-km mutant construction
The invA (BMEI0215) 1.6-kb fragment was PCR amplified using the 466 (5′-AGGATGTGAC CCGTTTCGAT-3′) and 467 (3′-CGATGCGCAA AATGATAAGG-5′) oligonucleotides. The amplification product was directly cloned into the pCR2.1-TOPO plasmid (Invitrogen-Life Technologies, K4500-01) to generate the pJA1 plasmid. The pJA1 plasmid was digested at the ClaI unique site to interrupt the invA gene and introduce the kanamycin resistant cassette, thereby generating plasmid pJA2. The kanamycin resistance gene (1100 pb) was obtained by PCR from the plasmid pSU4111. The ClaI restriction site in the kanamycin gene was inserted by PCR. The generated pJA2 plasmid was digested with the EcoRI enzyme to obtain the 2.9-kb fragment containing the invA gene interrupted by the kanamycin cassette. We cloned the resulting fragment into the suicide plasmid pKOK.4 to generate the plasmid pJA3. The pJA3 plasmid was introduced into B. melitensis 133 from E. coli S17.1 through conjugation assays, and the trans-conjugated colonies were chosen from plates containing kanamycin, tetracycline and ampicillin (all antibiotics from Sigma Aldrich; tetracycline, T7660). Kanamycin-resistant colonies susceptible to tetracycline and ampicillin were chosen. Interruption of the invA gene was confirmed through PCR and sequencing.
Complementation of the B. melitensis 133 invA-km mutant strain
To restore the invA gene, the plasmid pBBR4MCS was used. The complete invA gene contained in the 1.6-kb amplicon was subcloned into the pBBR4MCS-EcoRI-digested plasmid to generate plasmid pJA4. The electrocompetent B. melitensis 133 invA-km mutant was electroporated with the pJA4 plasmid. The electroporation conditions were as follows: 1.6 kV voltage, 50 μF capacitance, and less than 200 Ω resistance with 4 to 8 ms of electroporation time. Trans-conjugating colonies were chosen from plates containing ampicillin and kanamycin. The presence of the plasmid was determined through alkaline lysis, and analysis was performed via PCR.
HeLa cell invasion
HeLa cells were used as a model to identify Brucella invasion. These epithelial-like cells were grown at 37 °C in a 5% CO2 atmosphere in Dulbecco’s minimal essential medium (DMEM, Invitrogen-Life Technologies, 11995-065) supplemented with 10% fetal bovine serum (Invitrogen-Life Technologies,1 12483-020), 2 mmol/L glutamine (Invitrogen-Life Technologies, 25030-081), and 1.1% MEM non-essential amino acids (Invitrogen-Life Technologies, 11140-50). Cells were seeded (5 × 105) in 24-well cell culture plates one day before each infection assay. HeLa cells were infected as previously described,Citation35 with different B. melitensis strains at a multiplicity of infection (MOI) of 500 bacteria per cell. The culture plates were centrifuged for 10 min at 400 g at room temperature and placed in an incubator with a 5% CO2 atmosphere at 37 °C. After 15 min, which was considered time zero (incubation point 0 h), the wells were washed five times with phosphate-buffered saline (PBS) solution, and the monolayers were further incubated with cell culture medium supplemented with 100 μg of gentamicin (Sigma Aldrich, G1272) per mL for 60 min (or less for shorter time periods) to kill extracellular bacteria. The infected cells were washed three times with PBS and lysed with 0.2% Triton X-100 (Sigma Aldrich, T8787) at 30 min, 60 min, and 120 min pi. For bacterial counting, serial dilutions were performed and plated on Brucella agar to determine the colony-forming units (CFU). Experiments were performed in triplicate and repeated three times.
Phagocytosis and intracellular survival in macrophages
For the phagocytosis and intracellular survival assay, we used primary cultures of goat macrophages, as these cells are the principal target of B. melitensis. Blood-derived caprine macrophages were cultured in RPMI 1640 (Invitrogen-Life Technologies, 23400-062) supplemented with 2.2 mmol/L l-glutamine, 1.1% MEM non-essential amino acids, 1.1 mmol/L sodium pyruvate (Invitrogen-Life Technologies, 11360-070), 7.5% sodium bicarbonate (Sigma Aldrich, S5761), and 10% autologous serum (CRPMI). For isolation of caprine macrophages, blood was collected by jugular venipuncture (average: 200 mL) and re-suspended in 8 mL of ACD (74.8 mmol/L sodium citrate tribasic; S4641), 38 mmol/L citric acid (251275), and 135.9 mmol/L dextrose (D9559, all reagents from Sigma Aldrich). The blood was obtained from healthy, two-year-old brucellosis-free goats. The animal protocols and management were approved by the Internal Committee for Laboratory Animal Welfare of the National Autonomous University of Mexico. The blood was centrifuged at 1000 g for 20 min to obtain the leukocyte-rich fraction. This fraction was re-suspended in PBS-citric acid in a total volume of 30 mL. A Percoll® (GE Healthcare, 17089101) gradient of 1.074 g/mL was layered in a microfuge tube, and the leukocyte fraction was gently poured. Isopycnic centrifugation was performed at 1000 g for 30 min for leukocyte separation. The cells were seeded in 75-cm2 tissue culture flasks, and monocytes were separated via their adherent capacity. To verify monocyte to macrophage differentiation (13 d of incubation), nitric oxide (NO2) production was measured with the Griess reagent (Promega, G2930), according to the manufacturer’s instructions. For macrophage infection, 5 × 104 caprine macrophages were seeded on 24-well cell culture dishes one day before infection. Macrophages were infected as previously describedCitation36 with different B. melitensis strains at an MOI of 100 bacteria per cell. Intracellular bacteria were determined in the same manner as the invasion assay. Bacterial concentrations were determined at the beginning of the infection and at 4 h, 24 h, and 48 h pi. Experiments were performed in triplicate and repeated three times.
Macrophage immunofluorescence
The intracellular trafficking of B. melitensis strains was analyzed by infecting J774A.1 murine macrophage cells in the same manner described for goat macrophages. These cells were chosen because they are an accurate model for intracellular trafficking analysis.Citation37,Citation38 For immunofluorescence microscopy, infected J774A.1 cells were seeded on circular glass coverslips. The cells were fixed with 3% paraformaldehyde (Sigma Aldrich, P6148) for 15 min or with cool methanol (Sigma Aldrich, 322415) for 4 min. The cells were PBS-washed three times, and free radicals were neutralized with 50 mmol/L PBS-NH4Cl (Sigma Aldrich, 254134), only in paraformaldehyde-fixed cells. Incubation in 0.1% PBS-saponin for 1 min and 0.1% PBS-Triton for 4 min was performed before blocking the free receptors with PBS-10% horse serum. Primary antibody incubation was performed with goat polyclonal anti-EEA1 (Santacruz Biotechnology, SC6415), goat polyclonal anti-calregulin (Santacruz, T-19 sc-7431), mouse monoclonal anti-(H43A) LAMP1 (ABCAM, AB25630), goat polyclonal anti-cathepsin D (Santacruz, G-19 sc-6494), goat polyclonal anti-Brucella and mouse polyclonal anti-Brucella (both antibodies from our laboratory stock, Brucellosis Laboratory, College of Veterinary Medicine, UNAM). All incubations were performed for 40 min. Fluorescent secondary antibodies (Alexa 594, A11005, and Alexa 488, A1100; both reagents from Invitrogen-Life Technologies) were incubated for 40 min after the initial primary antibody incubation. Finally, coverslips were washed and mounted on glass slides with Mowiol® (Sigma Aldrich, 324590). Colocalization analysis was performed with a Leica DM100 immunofluorescence microscope (Leica Microsystems), and microphotographs were taken with an Olympus FV100 confocal microscope (Olympus America Corporate).
Infection of BALB/c mice
BALB/c female mice that were 6- to 8-wk-old were used and distributed homogeneously among 5 independent groups. Each group had 17 mice, and which were inoculated intraperitoneally with 0.1 mL of PBS containing 104 UFC of each bacterial strainCitation39 or 0.1 mL of PBS only (control group). Sampling was performed at 7, 15, 30, 60, and 90 d pi by sacrificing 3 mice per bacterial group. All mouse maintenance and procedures were approved by the Internal Committee for Laboratory Animal Welfare of the National Autonomous University of Mexico following ethical international standards.
Residual virulence in the murine model
At each sampling time point, the spleens were aseptically collected. The weight of each spleen was measured and recorded. Later, the spleens were macerated in 1 mL of PBS to determine the bacterial concentration and CFU counts, which were obtained with 10-fold serial dilutions and agar plating.
Humoral immune response
Sera obtained during the different sampling periods were collected for antibody isotype determination by ELISA. The samples were taken from whole blood obtained from the orbital sinus of desensitized mice. IgG1, IgG2a, IgG2b, IgG3, and IgM antibodies were analyzed with Mouse Monoclonal Antibody Isotyping Reagents (Sigma Aldrich, ISO2-1) following the manufacturer’s instructions. Briefly, ELISA plates were prepared with 100 ng of B. melitensis LPS (laboratory stock, Brucellosis Laboratory, College of Veterinary Medicine, UNAM). The plates were maintained at 4 °C before the ELISA procedure. PBS-5% skim milk was used to block unspecific receptors, and plates were incubated for 1 h at 37 °C. Next, the plates were washed three times with 0.05% PBS-Tween 20 (Sigma Aldrich, P9416). Sera were diluted to a working concentration of 1:200, and samples were analyzed in triplicate; 100 μL of each diluted sample was deposited in a well of a prepared ELISA plate. The ELISA plates were incubated at 37 °C for 1 h and washed three times with 0.05% PBS-Tween 20. Before adding 100 μL of mouse anti-IgG or anti-IgM antibody isotypes and incubating for 30 min at room temperature, every mouse anti-IgG or anti-IgM antibody was diluted 1:1000. Later, the plates were washed as previously described. The rabbit anti-goat IgG peroxidase-labeled (Sigma Aldrich, A5420) antibody was diluted 1:5000 before depositing 100 μL in each well and incubating at room temperature for 15 min. The plates were washed as previously described. The enzyme substrate was prepared as follows. A 1 mg/mL solution of 5-aminosalicylic acid (Sigma Aldrich, A3537) in 0.02 M sodium phosphate (Sigma Aldrich, 342483) at pH 6.8 was prepared. One hundred microliters of 1% H2O2 (Sigma Aldrich, 216763) was added for every 10 mL of fresh substrate solution. The substrate solution was added to each well (100 μL) and incubated in the dark for 15 min at room temperature. Then stop solution (50 μL of 3 N NaOH, Sigma Aldrich, S5881) was added to each well. Absorbance measurements at 490 nm were performed with a VECTOR3 multilabel counter (PerkinElmer) with negative controls.
IFN-γ transcript quantification by RT-PCR
To provide evidence for the Th1 immune response IFN-γ, transcription expression was measured with real-time PCR (RT-PCR). RNA was obtained from whole blood from infected mice at different time points. RNA isolation was performed with Tripure Isolation Reagent (Roche Diagnostics GmbH, 11667157001) following the manufacturer’s instructions. Total genomic DNA was eliminated by incubation with DNase I (Invitrogen-Life Technologies,18068-015). The RNA concentration and purity were determined by spectrophotometry. The Omniscript RT Kit (Qiagen, 20511) and Fermentas Oligo-dT 18 (Thermo Scientific, SO132) were used to transform mRNA (mRNA) into cDNA (cDNA). Reverse transcription was performed at 37 °C for 60 min followed by 95 °C for 5 min. The purity and concentration of the cDNA were determined by spectrophotometry. RT-PCR was performed with LightCycler 480 DNA SYBR Green Master I mix (Roche, 04707516001) using 100 ng of cDNA, 0.5 pM of oligonucleotides, and 8 μL of SyBR Green Master I mix in a final volume of 20 μL of nuclease-free water. The following oligonucleotide sequences were used: 5′-TGGAATCCTT GGCATCCATG AAAC-3′ and 5′-TAAAACGCAG CTCAGTAACA GTCCG-3′ for the β-actin transcript; 5′-AGCGGCTGAC TGAACTCAGA TTGTAG-3′ and 5′-GTCACAGTTT TCAGCTGTAT AGGG-3′ for the IFN-γ transcript.Citation40 RT-PCR was performed on a LightCycler 480 thermal cycler (Roche Diagnostics GmbH) following the manufacturer’s recommendations. The mRNA levels were normalized against mouse β-actin, and the ratio between infected and non-infected mice (control group) was determined at different times pi.
Statistical analysis
Statistical significance was calculated using ANOVA two-way analyses, and treatment mean comparisons were evaluated with the Bonferroni post-test analysis. For colocalization results, the geometric mean was used for ANOVA two-way analyses. The software used for these analyses was GraphPad Prism 5. The significance level was set at P < 0.05 or P < 0.001.
| Abbreviations: | ||
| NUDIX | = | (di)nucleoside oligophosphate molecules linked to other “X” molecules |
| pi | = | post-infection |
| BCV | = | Brucella-containing-vacuole |
| Bm | = | Brucella melitensis |
| CFU | = | colony-forming units |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by “Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT), Universidad Nacional Autónoma de México” Projects IN207608 and IT222511. J.A.P. received scholarships from CONACYT and PAPIIT.
References
- Corbel MJ. Brucellosis: an overview. Emerg Infect Dis 1997; 3:213 - 21; http://dx.doi.org/10.3201/eid0302.970219; PMID: 9204307
- Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, Fretin D, Whatmore AM, Cloeckaert A, Blasco JM, Moriyon I, et al. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med 2011; 102:118 - 31; http://dx.doi.org/10.1016/j.prevetmed.2011.04.007; PMID: 21571380
- Barbier T, Nicolas C, Letesson JJ. Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett 2011; 585:2929 - 34; http://dx.doi.org/10.1016/j.febslet.2011.08.011; PMID: 21864534
- Rossetti CA, Galindo CL, Garner HR, Adams LG. Transcriptional profile of the intracellular pathogen Brucella melitensis following HeLa cells infection. Microb Pathog 2011; 51:338 - 44; http://dx.doi.org/10.1016/j.micpath.2011.07.006; PMID: 21798337
- Mirabella A, Terwagne M, Zygmunt MS, Cloeckaert A, De Bolle X, Letesson JJ. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J Bacteriol 2013; 195:453 - 65; http://dx.doi.org/10.1128/JB.01336-12; PMID: 23161025
- Teixeira-Gomes AP, Cloeckaert A, Zygmunt MS. Characterization of heat, oxidative, and acid stress responses in Brucella melitensis.. Infect Immun 2000; 68:2954 - 61; http://dx.doi.org/10.1128/IAI.68.5.2954-2961.2000; PMID: 10768994
- Roop RM 2nd, Gee JM, Robertson GT, Richardson JM, Ng W-L, Winkler ME. Brucella stationary-phase gene expression and virulence. Annu Rev Microbiol 2003; 57:57 - 76; http://dx.doi.org/10.1146/annurev.micro.57.030502.090803; PMID: 12730323
- Dozot M, Boigegrain RA, Delrue RM, Hallez R, Ouahrani-Bettache S, Danese I, Letesson JJ, De Bolle X, Köhler S. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB.. Cell Microbiol 2006; 8:1791 - 802; http://dx.doi.org/10.1111/j.1462-5822.2006.00749.x; PMID: 16803581
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 2010; 74:171 - 99; http://dx.doi.org/10.1128/MMBR.00046-09; PMID: 20508246
- Guranowski A. Analogs of diadenosine tetraphosphate (Ap4A). Acta Biochim Pol 2003; 50:947 - 72; PMID: 14739989
- Pietrowska-Borek M, Nuc K, Zielezińska M, Guranowski A. Diadenosine polyphosphates (Ap3A and Ap4A) behave as alarmones triggering the synthesis of enzymes of the phenylpropanoid pathway in Arabidopsis thaliana.. FEBS Open Bio 2011; 1:1 - 6; http://dx.doi.org/10.1016/j.fob.2011.10.002; PMID: 23650569
- Lee PC, Bochner BR, Ames BN. AppppA, heat-shock stress, and cell oxidation. Proc Natl Acad Sci U S A 1983; 80:7496 - 500; http://dx.doi.org/10.1073/pnas.80.24.7496; PMID: 6369319
- Bochner BR, Lee PC, Wilson SW, Cutler CW, Ames BN. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 1984; 37:225 - 32; http://dx.doi.org/10.1016/0092-8674(84)90318-0; PMID: 6373012
- McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci 2006; 63:123 - 43; http://dx.doi.org/10.1007/s00018-005-5386-7; PMID: 16378245
- Gaywee J, Xu W, Radulovic S, Bessman MJ, Azad AF. The Rickettsia prowazekii invasion gene homolog (invA) encodes a Nudix hydrolase active on adenosine (5′)-pentaphospho-(5′)-adenosine. Mol Cell Proteomics 2002; 1:179 - 85; http://dx.doi.org/10.1074/mcp.M100030-MCP200; PMID: 12096117
- Bessman MJ, Walsh JD, Dunn CA, Swaminathan J, Weldon JE, Shen J. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a Nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)-adenosine (Ap5A). J Biol Chem 2001; 276:37834 - 8; PMID: 11479323
- Edelstein PH, Hu B, Shinzato T, Edelstein MA, Xu W, Bessman MJ. Legionella pneumophila NudA Is a Nudix hydrolase and virulence factor. Infect Immun 2005; 73:6567 - 76; http://dx.doi.org/10.1128/IAI.73.10.6567-6576.2005; PMID: 16177332
- Ismail TM, Hart CA, McLennan AG. Regulation of dinucleoside polyphosphate pools by the YgdP and ApaH hydrolases is essential for the ability of Salmonella enterica serovar typhimurium to invade cultured mammalian cells. J Biol Chem 2003; 278:32602 - 7; http://dx.doi.org/10.1074/jbc.M305994200; PMID: 12824172
- Cartwright JL, Britton P, Minnick MF, McLennan AG. The IalA invasion gene of Bartonella bacilliformis encodes a (de)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem Biophys Res Commun 1999; 256:474 - 9; http://dx.doi.org/10.1006/bbrc.1999.0354; PMID: 10080922
- Rajashekara G, Glover DA, Banai M, O’Callaghan D, Splitter GA. Attenuated bioluminescent Brucella melitensis mutants GR019 (virB4), GR024 (galE), and GR026 (BMEI1090-BMEI1091) confer protection in mice. Infect Immun 2006; 74:2925 - 36; http://dx.doi.org/10.1128/IAI.74.5.2925-2936.2006; PMID: 16622231
- Jiang X, Baldwin CL. Effects of cytokines on intracellular growth of Brucella abortus.. Infect Immun 1993; 61:124 - 34; PMID: 8418034
- Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol 2012; 33:397 - 405; http://dx.doi.org/10.1016/j.it.2012.03.003; PMID: 22560866
- Delrue RM, Martinez-Lorenzo M, Lestrate P, Danese I, Bielarz V, Mertens P, De Bolle X, Tibor A, Gorvel JP, Letesson JJ. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol 2001; 3:487 - 97; http://dx.doi.org/10.1046/j.1462-5822.2001.00131.x; PMID: 11437834
- von Bargen K, Gorvel JP, Salcedo SP. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Rev 2012; 36:533 - 62; http://dx.doi.org/10.1111/j.1574-6976.2012.00334.x; PMID: 22373010
- Starr T, Child R, Wehrly TD, Hansen B, Hwang S, López-Otin C, Virgin HW, Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 2012; 11:33 - 45; http://dx.doi.org/10.1016/j.chom.2011.12.002; PMID: 22264511
- Baldwin CL, Goenka R. Host immune responses to the intracellular bacteria Brucella: does the bacteria instruct the host to facilitate chronic infection?. Crit Rev Immunol 2006; 26:407 - 42; http://dx.doi.org/10.1615/CritRevImmunol.v26.i5.30; PMID: 17341186
- Lavigne JP, Patey G, Sangari FJ, Bourg G, Ramuz M, O’Callaghan D, Michaux-Charachon S. Identification of a new virulence factor, BvfA, in Brucella suis. Infect Immun 2005; 73:5524 - 9; http://dx.doi.org/10.1128/IAI.73.9.5524-5529.2005; PMID: 16113268
- Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys 2005; 433:129 - 43; http://dx.doi.org/10.1016/j.abb.2004.08.017; PMID: 15581572
- Kim S, Kurokawa D, Watanabe K, Makino S, Shirahata T, Watarai M. Brucella abortus nicotinamidase (PncA) contributes to its intracellular replication and infectivity in mice. FEMS Microbiol Lett 2004; 234:289 - 95; http://dx.doi.org/10.1111/j.1574-6968.2004.tb09546.x; PMID: 15135535
- Del Giudice MG, Ugalde JE, Czibener C. A lysozyme-like protein in Brucella abortus is involved in the early stages of intracellular replication. Infect Immun 2013; 81:956 - 64; http://dx.doi.org/10.1128/IAI.01158-12; PMID: 23319555
- Murphy EA, Parent M, Sathiyaseelan J, Jiang X, Baldwin CL. Immune control of Brucella abortus 2308 infections in BALB/c mice. FEMS Immunol Med Microbiol 2001; 32:85 - 8; http://dx.doi.org/10.1111/j.1574-695X.2001.tb00536.x; PMID: 11750226
- Zhang X, Ren J, Li N, Liu W, Wu Q. Disruption of the BMEI0066 gene attenuates the virulence of Brucella melitensis and decreases its stress tolerance. Int J Biol Sci 2009; 5:570 - 7; http://dx.doi.org/10.7150/ijbs.5.570; PMID: 19742243
- Caro-Hernández P, Fernández-Lago L, de Miguel MJ, Martín-Martín AI, Cloeckaert A, Grilló MJ, Vizcaíno N. Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis.. Infect Immun 2007; 75:4050 - 61; http://dx.doi.org/10.1128/IAI.00486-07; PMID: 17562767
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2001.
- Pizarro-Cerdá J, Moreno E, Sanguedolce V, Mege JL, Gorvel JP. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun 1998; 66:2387 - 92; PMID: 9573138
- Pei J, Wu Q, Kahl-McDonagh M, Ficht TA. Cytotoxicity in macrophages infected with rough Brucella mutants is type IV secretion system dependent. Infect Immun 2008; 76:30 - 7; http://dx.doi.org/10.1128/IAI.00379-07; PMID: 17938217
- Arenas GN, Staskevich AS, Aballay A, Mayorga LS. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect Immun 2000; 68:4255 - 63; http://dx.doi.org/10.1128/IAI.68.7.4255-4263.2000; PMID: 10858243
- Naroeni A, Jouy N, Ouahrani-Bettache S, Liautard JP, Porte F. Brucella suis-impaired specific recognition of phagosomes by lysosomes due to phagosomal membrane modifications. Infect Immun 2001; 69:486 - 93; http://dx.doi.org/10.1128/IAI.69.1.486-493.2001; PMID: 11119541
- Fretin D, Fauconnier A, Köhler S, Halling S, Léonard S, Nijskens C, Ferooz J, Lestrate P, Delrue RM, Danese I, et al. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol 2005; 7:687 - 98; http://dx.doi.org/10.1111/j.1462-5822.2005.00502.x; PMID: 15839898
- Murray LJ, Lee R, Martens C. In vivo cytokine gene expression in T cell subsets of the autoimmune MRL/Mp-lpr/lpr mouse. Eur J Immunol 1990; 20:163 - 70; http://dx.doi.org/10.1002/eji.1830200124; PMID: 1968391
- Hernández-Castro R, Verdugo-Rodríguez A, Gutiérrez-Pabello JA, Adams LG, Suárez-Güemes F, Sahagún-Ruíz A. Identification of four genes of the Brucella melitensis ATP synthase operon F0 sector: relationship with the Rhodospirillaceae family. Microb Comp Genomics 2000; 5:163 - 71; http://dx.doi.org/10.1089/omi.1.2000.5.163; PMID: 11252353
- Castañeda-Ramírez A, Puente JL, González-Noriega A, Verdugo-Rodríguez A. Silencing of VAMP3 expression does not affect Brucella melitensis infection in mouse macrophages. Virulence 2012; 3:434 - 9; http://dx.doi.org/10.4161/viru.21251; PMID: 23076244
- Moncalián G, Grandoso G, Llosa M, de la Cruz F. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J Mol Biol 1997; 270:188 - 200; http://dx.doi.org/10.1006/jmbi.1997.1082; PMID: 9236121
- Kokotek W, Lotz W. Construction of a mobilizable cloning vector for site-directed mutagenesis of gram-negative bacteria: application to Rhizobium leguminosarum.. Gene 1991; 98:7 - 13; http://dx.doi.org/10.1016/0378-1119(91)90097-U; PMID: 2013412
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995; 166:175 - 6; http://dx.doi.org/10.1016/0378-1119(95)00584-1; PMID: 8529885
