Abstract
Plant resistance (R) proteins perceive specific pathogen effectors from diverse plant pathogens to initiate defense responses, designated effector-triggered immunity (ETI). Plant R proteins are mostly nucleotide binding-leucine rich repeat (NB-LRR) proteins, which recognize pathogen effectors directly or indirectly through sophisticated mechanisms. Upon activation by effector proteins, R proteins elicit robust defense responses, including a rapid burst of reactive oxygen species (ROS), induced biosynthesis and accumulation of salicylic acid (SA), a rapid programmed cell death (PCD) called hypersensitive response (HR) at the infection sites, and increased expression of pathogenesis-related (PR) genes. Initiation of ETI is correlated with a complex network of defense signaling pathways, resulting in defensive cellular responses and large-scale transcriptional reprogramming events. In this review, we highlight important recent advances on the recognition of effectors, regulation and activation of plant R proteins, dynamic intracellular trafficking of R proteins, induction of cell death, and transcriptional reprogramming associated with ETI. Current knowledge gaps and future research directions are also discussed in this review.
Introduction
All organisms constantly interact with other organisms and the surrounding environment. Because of their sessile nature, plants cannot escape harsh environments or detrimental organisms. However, partially due to their rigid cell wall, plants are competent in dealing with a wide variety of abiotic and biotic stresses.Citation1,Citation2 One of the major challenges that plants face is defending against many pathogens that cause diseases in plants.Citation3 Unlike animals, which have developed a circulatory system and specialized immune cells, plants rely on both conserved and unique defense mechanisms to defend themselves against pathogen infection.
The first layer of plant defense is a conserved defense mechanism shared by plants and animals. Plants and animals both have developed the ability to detect conserved small molecules associated with microbes called MAMPs, or microbe-associated molecular patterns. This first layer of immunity is called MAMP-triggered immunity (MTI). Two major MAMPs associated with plant bacterial pathogens, flagellin and EF-Tu, are recognized by the pattern recognition receptors (PRRs), FLS2 kinase and EFR kinase, respectively. Chitin is believed to be the major MAMP from plant fungal pathogens and the receptor for chitin has been shown to be a LysM domain receptor-like kinase, CERK1Citation4 or RLK1,Citation5 in Arabidopsis. The consequence of this recognition includes activation of a Mitogen-activated protein kinase (MAPK) pathway, oxidative burst, callose deposition, and restriction of pathogen growth, etc.Citation6 In order to cause diseases, plant pathogens such as fungi, oomycetes, and bacteria deliver effectors into plant cells. These effectors collectively suppress MTI; therefore, the pathogens carrying these effectors cause diseases on their host plants. During plant–pathogen coevolution, plants have developed resistance (R) proteins to recognize at least some of the effectors and activate plant defense. Effectors that are recognized by corresponding R proteins are called avirulence (Avr) proteins.Citation7 A pathogen that carries an Avr protein, which triggers plant resistance, is called an avirulent pathogen. Unlike virulent pathogens, which cause diseases, avirulent pathogens trigger plant defense. Infection by an avirulent pathogen will produce rapid programmed cell death (PCD), called the hypersensitive response (HR), in the local tissue.Citation8 Meanwhile, plants undergo transcriptional reprogramming and the expression of plant defense genes is activated.Citation9,Citation10 This second layer defense is called effector-triggered immunity (ETI). Recognition of avirulent pathogens will also generate immune signal plant hormone salicylic acid (SA) and activate systemic acquired resistance (SAR) in the whole plant.Citation11,Citation12 SAR is a broad-spectrum, long-lasting resistance in the whole plant after local exposure to an avirulent or virulent pathogen.
This review will be focused on ETI. In the last decade, tremendous progress has been made toward a better understanding of the molecular mechanism of ETI. Here we describe in detail the recent exciting developments on how plants deploy ETI to combat pathogens. The current knowledge gaps and future directions will be discussed as well.
R Proteins
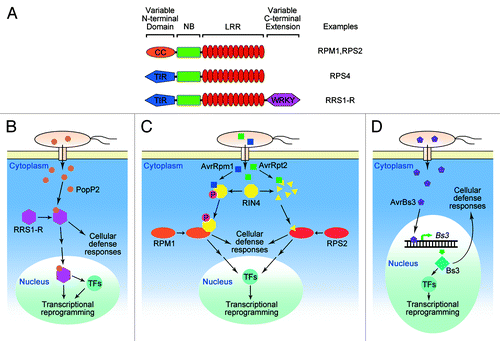
The majority of R proteins are intracellular nucleotide binding-leucine rich repeat (NB-LRR) proteins (), which are subclassified based on their variable N-terminal domain into coiled-coil (CC)-NB-LRR or toll/interleukin 1 receptor-like (TIR)-NB-LRR protein families. Some R proteins have a C-terminal extension. For example, Arabidopsis RRS1-R (resistance to Ralstonia solanacearum-resistant allele), includes a WRKY transcription factor domain following the LRR at the C-terminal (). Inactive R protein binds ADP, with intramolecular interactions between its different domains.Citation13 Upon induction, R proteins undergo conformational change, which releases the hidden nucleotide-binding site from intramolecular interactions in the presence of effector ligands, leading to ADP/ATP nucleotide exchange and hydrolysis in the central NBS domain.Citation13,Citation14 It is generally believed that ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) is required for the disease resistance mediated by the TIR-NB-LRR class R proteins, while NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1) plays an essential role in the activation of resistance signaling mediated by the CC-NB-LRR class of resistance proteins.Citation15
Figure 1. Recognition of pathogen effectors by plant resistance (R) proteins. (A) Structures of R proteins. The majority of R proteins are NB (nucleotide binding)-LRR (leucine-rich repeat) proteins, which are grouped into CC (coiled-coil)-NB-LRRs and TIR (toll/interleukin 1 receptor like)-NB-LRRs based on their distinct N-terminal domains. Some R proteins have a C-terminal extension required for their biological function, such as RRS1-R, which has a WRKY domain. (B–D) Simplified model showing examples of recognition between plant R proteins and pathogen effectors. (B) RRS1-R recognizes effector PopP2 through direct interaction, and relocated into nucleus to promote defense-related gene expression through its C-terminal WRKY transcription activator domain. (C) RIN4 is targeted by type III effectors AvrRpm1 and AvrRpt2, and surveilled by resistance proteins RPM1 and RPS2, respectively. RPM1 recognizes AvrRpm1-meidtaed phosphorylation of RIN4, while RPS2 detects the degradation of RIN4 by cysteine protease activity of AvrRpt2. (D) TAL (Transcription activator-like) effector AvrBs3 from xanthomonads (Xanthomonas spp.) induces the expression of R protein Bs3 after its nuclear localization to activate ETI (effector-triggered immunity). Recognition of effectors by plant R proteins triggers robust cellular defense responses, and transcriptional reprogramming of defense genes. TFs, transcription factors.
In response to pathogen infections, R proteins induce dramatic local physiological responses, including oxidative burst, production and accumulation of SA and nitric oxide (NO), and expression of pathogenesis-related (PR) genes and PCD, at the cost of altering normal physiological processes. Therefore, the activity of R proteins needs to be tightly controlled under uninfected conditions.
Regulation of R Genes
Some R genes only express in response to Avr effectors.Citation16-Citation18 Rice (Oryza sativa) R gene Xa27 is induced when challenged by Xanthomonas oryzae pv oryaze (Xoo) carrying avrXa27 effector gene.Citation19 AvrXa27 belongs to the AvrBs3/PthA TAL effector family, the members of which contain repeats of 34 amino-acids with DNA-binding activity and a conserved C-terminal region with nuclear localization signal (NLS) motifs and transcription activation domains.Citation20 AvrXa27 differs from other members in the same family based on the number and arrangement of the repeats, which contribute to target binding specificity.Citation19 The expression of Xa27 is only detectable upon inoculation of the Xoo strain that delivers AvrXa27, while undetectable when challenged with PXO99AME1 strain, which has an insertion mutation in the avrXa27 gene.Citation19
RNA silencing is a central regulator that controls gene expression at the transcriptional level via DNA methylation (TGS, transcriptional gene silencing) and posttranscriptional level via direct mRNA interference mediated by small RNAs (PTGS, posttranscriptional gene silencing).Citation21 Recently, RNA silencing has appeared as a key regulatory mechanism in negatively regulating expression of a cluster of R genes.Citation22 It was first determined that several RPP5 locus R genes, including RPP4 and SNC1, are regulated by siRNA-mediated PTGS. Upregulation of SNC1 transcription is found in mutants, dcl4 and ago1, which abrogate RNA silencing. In addition, microRNA miR482 was shown to target many R genes in different species and subsequently cause the cleavage of R gene mRNA and production of secondary siRNAs.Citation23 More recently, it has been demonstrated that RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) and miR472 play important roles in ETI and basal defenses through PTGS of a subset of R genes.Citation24 Mutants of rdr6 and miR472m displayed enhanced RPS5-mediated resistance with high accumulation of RPS5 transcripts, suggesting that both RDR6 and miR472 act as negative regulators of ETI through post-transcriptional control of disease resistance genes.
Regulation of R Protein Level and Stability
Many R genes are expressed constitutively under uninfected conditions. Regulation of R protein level and stability is another mechanism for plants to control R protein signaling, avoiding unnecessary activation of defense responses and autoimmunity. A chaperone protein complex containing HSP90, SGT1, and RAR1 contributes to the stability and proper folding of R proteins during activation,Citation25 leading to downstream signaling. In addition to its positive regulatory role as a chaperone for R proteins, SGT1b was also demonstrated to negatively regulate the accumulation of RPS5 and SNC1.Citation26,Citation27 An effort to screen for mutants with constitutive defense responses in npr1–1 background identified a tetratricopeptide repeat-domain containing protein-encoding gene, SUPPRESSOR OF rps4-RLD, known as SRFR1. SRFR1 functions together with SGT1 to negatively regulate the accumulation of SNC1, RPS2, and RPS4 to prevent auto-activation of plant immunity.Citation27 Further study showed that F-box protein CPR1/CPR30 targets SNC1 and RPS2 for degradation through the SKP1-Cullin1-F-box (SCF) E3 ubiquitin ligase complex.Citation28,Citation29 Light has been demonstrated to regulate plant R protein-mediated resistance and photoreceptors are involved in this regulation.Citation30-Citation34 In-depth study showed that blue light receptors, CRY2 and PHOT2, stabilize R protein HRT against Turnip Crinkle Virus (TCV) infection by suppressing COP1 E3 ubiquitin ligase-mediated degradation of HRT.Citation34 R proteins stay inactive and are tightly controlled under non-pathogenic conditions even though they are required for plants to initiate robust defense responses to fight back pathogen attacks.
The Recognition between R Proteins and Pathogen Effectors
The first step for plants to activate an effective ETI upon infection from pathogens carrying avirulence genes is to recognize those Avr effectors that have been delivered into plant cells. Effector proteins usually interfere with plant basal resistance, or MTI, to help pathogens overcome plant resistance. Plants have evolved distinct mechanisms to recognize effectors. The direct interaction between a plant R protein and an Avr effector protein can be demonstrated by a recent example in Arabidopsis, where R protein RRS1-R recognizes effector protein PopP2 from a causal agent of bacterial wilt, R. solanacearum (). Molecular evidence was provided to show the direct protein-protein interactions between RRS1-R and PopP2 in Arabidopsis-resistant Nd-1 ecotype,Citation35 and RRS1-R was shown to translocate into nucleus after PopP2 binding to regulate downstream gene expression (). Direct recognition was also shown by the recognition of fungal effector AvrPita from Magnaporthe grisea by the rice R protein Pi-ta to trigger a signal transduction cascade that led to resistance.Citation36 AvrPita is predicated to encode a neutral zinc metalloprotease.Citation37 A variant of AvrPita, AvrPita176, which lacks the N-terminal secretory signal and pro-protein sequences of AvrPita, elicited Pi-ta-dependent resistance responses when transiently expressed inside rice cells. Yeast two-hybrid and in vitro binding assays showed direct interaction between AvrPita176 and the Pi-ta leucine-rich repeat domain.Citation36 A recent study showed direct recognition of multiple effectors by the same R protein, which usually happens through indirect recognition. Evidence was provided to show that two distinct effectors, Avr1-CO39 and AvrPia, from rice blast fungal pathogen M. oryzae interacted directly with rice R protein RGA5, which worked in pair with RGA4 to induce resistance.Citation38 However, only a few cases of direct recognition between an R protein and an Avr effector have been demonstrated, even though direct recognition possibly contributes to gene-specific evolution diversity of both R and Avr genes, while indirect recognition imposes selection against the biological function of Avr effectors.Citation39
The guard model has been developed to explain indirect recognition between R proteins and Avr effectors in plant, in which a target host protein is manipulated by pathogen effectors and the perturbation of the target protein is monitored by plant R proteins. Plant RPM1-INTERACTING PROTEIN 4 (RIN4) has been shown to play a crucial role in the recognition of AvrRpm1 and AvrRpt2 by the corresponding plant R proteins, RPM1 and RPS2, respectivelyCitation40,Citation41 (). RPM1 perceives the phosphorylation of RIN4, while RPS2 senses the disappearance of RIN4 protein.Citation40,Citation41 Recent studies showed that a receptor-like cytoplasmic kinase, RIPK (RPM1-induced protein kinase), physically interacts with RIN4 and phosphorylates RIN4 at multiple amino acid residues in response to bacterial effectors AvrRpm1 and AvrB.Citation42,Citation43 AvrRpt2 induces RIN4 degradation with its cysteine protease activityCitation44-Citation46 (). RIN4 is a regulator of plant MTI. RIN4 controls stomatal apertures through associated plasma membrane H(+)-ATPases, AHA1 and AHA2, to resist pathogen invasion.Citation47,Citation48 Independently evolved virulence effectors targeting hub proteins, such as RIN4, in plant immune system networks enable the plant to respond to infections by monitoring these hub proteins with a relatively limited number of R proteins.Citation49,Citation50 Another example of a guarded hub protein is EDS1, which was assumed to constitute a common downstream signaling component of several TIR-NBS-LRR proteins based on genetic studies.Citation51 EDS1 was recently shown to be a target for AvrRps4 and HopA1 effectors, and was guarded by RPS4, RPS6, and SNC1 resistance proteins.Citation52,Citation53
Some of the plant target proteins evolved into decoys, which were no longer biologically functional, but remained to be the targets for pathogen effectors and guardees for plant R proteins. ZED1 in Arabidopsis is a nonfunctional kinase that interacts with and is acetylated by HopZ1a, a type III effector from Pseudomonas syringae that triggers ZAR1-mediated defense responses. HopZ1a is an acetyltransferase that is hypothesized to target kinases to promote pathogen virulence. Conserved domain analysis of the protein sequence indicated that ZED1 is a kinase, however, showed no kinase activity in phosphorylation assays. ZED1 interacts with both HopZ1a and ZAR1, and is required for the recognition of HopZ1a by the ZAR1 to induce a robust immune response to limit bacteria growth. Thus, ZED1 very likely evolved as a decoy to trap HopZ1a and lure it to the ZAR1 resistance-complex.Citation54 Even though the decoy model is supported by multiple cases, it is arguable that discrimination between the decoy model and the guard model remains challenging and requires further experimental evidences.Citation55 An interesting case involves tomato (Solanum lycopersicum) protein kinase Pto, which is targeted by the corresponding Avr effector, AvrPto, from P. syringae pv tomato. It was first described as an example for direct recognition supported by evidence from the yeast two-hybrid, protein mutagenesis and infection assays that strongly indicated direct physical interaction between Pto and AvrPto.Citation56-Citation59 Further study showed the involvement of NBS-LRR protein Prf in AvrPto-Pto triggered immune response, leading to Pto kinase being considered as a guardee monitored by Prf resistance protein.Citation60 More recent work proposed that Pto acts as a decoy in competition with FLS2 for AvrPto binding.Citation61,Citation62 FLS2 is a receptor kinase which recognizes bacterial MAMP flagellin to trigger MTI and is believed to be the real target of AvrPto.Citation63
The knowledge of a unique way of recognition between a plant R protein and a pathogen Avr effector came from studies on transcription activator-like (TAL) effectors from Xanthomonas pathovars. These effectors enter the nucleus and bind to promoters to alter the expression of target genes in order to promote virulence. AvrBs3 from X. campestris pv vesicatoria targets the promoter of UPA20, which is a master regulator gene of cell enlargement in pepper, to help bacterial proliferation.Citation64 AvrBs3 is recognized by Bs3, a protein product of the R gene Bs3, which shares a similar promoter sequence to AvrBs3 target genes,Citation65,Citation66 and thus can be induced by AvrBs3 (). No Bs3 expression was detected in the absence of AvrBs3 in different plant tissues, indicating that Bs3 is not functional for normal cellular activities,Citation65,Citation66 and likely a decoy to trap AvrBs3 to initiate Bs3-mediated resistance.Citation67
Nuclear Function of R Proteins
Nuclear localization of several R proteins is essential for ETI, which leads to transcriptional activation of defense signaling. Deslandes et al. identified TIR-NB-LRR-WRKY protein RRS1-R to be the first R protein that has been shown to localize in the nucleus.Citation68 The presence of a C-terminal WRKY domain suggested a biological function for RRS1-R in the nucleus. Sequence analysis further showed that RRS1-R has an NLS signal.Citation68 Indeed, it has been shown that RRS1-R localized into the nucleus and its nuclear localization is dependent on the type III effector PopP2, which is recognized by the R protein RRS1-R.Citation35 RRS1-R was found to physically interact with its cognate effector PopP2 and to co-localize with PopP2 in the plant nucleus.Citation35 More clearly through quantitative fluorescence lifetime imaging, it is illustrated that barley (Hordeum vulgare) mildew R protein A10 (MLA10), recognizing effector AvrA10, traffics from the cytoplasm into the nucleus where its CC domain interacts with transcription factors HvWRKY1/2 and MYB6 in the nucleus to activate downstream transcriptional reprogramming.Citation69 P. syringae type III effector AvrRps4 is recognized by resistance protein RPS4 and triggers ETI. The cell has cytoplasmic and nucleoplasmic pools of EDS1, which must accumulate in the nucleus for RPS4 to trigger proper immunity processes.Citation52,Citation70 After injection into plant cells, AvrRps4 is cleaved to trigger RPS4-mediated signaling, which results in nuclear localization of RPS4 to interact with RRS1-R, a resistance receptor with a C-terminal WRKY transcription factor DNA-binding domain, to promote immunity.Citation71,Citation72 RPS4 is believed to be a receptor that intercepts targeting of EDS1 by effectors.Citation52 This interception and subsequent nuclear localization of RPS4-AvrRps4 triggers EDS1 to activate EDS1-dependent resistance genes, mounting an ETI response.
Trafficking of R Proteins between the Nucleus and Cytoplasm
Intracellular movement of different proteins between the cytoplasm and nucleus is an important feature in plant immunity.Citation73,Citation74 Recent studies revealed that nucleo-cytoplasmic trafficking though nuclear pore complexes plays an important role for R protein nuclear localization and the proper function of R proteins. In an effort to discover key components involved in R protein function, several mos (modifier of SNC1) mutants were identified in a suppressor screen in gain of function snc1 (Arabidopsis suppressor of npr1–1, constitutive 1) mutant, which showed a phenotype of constitutively activated plant defense due to a mutation in the TIR-NB-LRR R gene SNC1.Citation75 Among them, three MOS proteins that are important for nuclear pore function were required for the functions of R proteins. MOS3 is required for basal resistance and proper SNC1 function. mos3–1 mutant suppressed the constitutive activation of resistance phenotype in snc1 plants. Green fluorescent protein (GFP) tagging of MOS3 showed localization of MOS3-GFP to the nuclear envelope. Due to similarities in sequence between MOS3 and nucleoporin 96, it is likely that MOS3 encodes a nucleoporin that plays a central role in the nuclear transport of SNC1.Citation76 MOS6 encodes importin α3, which is instrumental in transport across the nuclear envelope. When MOS6 protein was tagged with GFP on the C-terminus, the GFP fluorescence signal was concentrated in the nucleus, supporting the nuclear localization pattern necessary for an importin protein. The mos6 mutant partially suppresses the constitutive defense phenotype in snc1 mutant plants, and mos6 mutant plants were more susceptible to a virulent oomycete pathogen.Citation77 MOS7 is homologous to nuclear pore complex component Nup88 in humans and Drosophila. MOS7 C-terminus GFP fusion protein showed accumulation at the nuclear rim. SNC1 has a nuclear localization sequence; however, in mos7–1 mutant, nuclear accumulation of SNC1 resistance protein is diminished.Citation78 NPR1 and EDS1 plant basal resistance proteins also have reduced localization to the nucleus in mos7 mutants.Citation78 Thus, MOS3, MOS6, and MOS7 have a central role in R protein nuclear localization.Citation78
Plant Defense Responses Associated with ETI
Localized programmed cell death
The striking characteristic of ETI is the hypersensitive response, which exhibits a rapid induction of programmed cell death at the local infection site (localized PCD). The primary purpose of this cell death is to kill biotrophic pathogens, which have to obtain nutrients from living cells. PCD has been classified into two different types, autolytic-PCD and nonautolytic-PCD,Citation79 with distinct characteristics. Autolytic-PCD involves the release of hydrolases from vacuoles, leading to a rapid cytoplasm clearance and localized cell death. Nonautolytic-PCD, however, does not release hydrolases that clear the cytoplasm even when the permeability of tonoplasts is persistently increased.Citation79 The cell death upon the activation of R protein signaling was described as a process of releasing vacuolar antibacterial proteins into the apoplast, resulting from the fusion of the central vacuole membrane and plasma membrane,Citation80 and thus is likely nonautolytic-PCD.Citation81
Regulation of localized PCD by salicylic acid concentration gradient and NPR proteins
PCD in Arabidopsis is under regulation by SA, NPR1, and SA receptors NPR3 and NPR4. Low level of SA suppresses cell death, while over-accumulation of SA induces cell death. When infected by an avirulent pathogen, the plants undergo PCD at the local infection sites. In an area up to 3.5 mm away from the center of cell death zone on Nicotiana tabacum L. leaves, the level of SA is 8-fold higher than in the zone 3.5–6.5 mm from the center 120 h post tobacco mosaic virus (TMV) infection, and 27-fold higher than in the zone 6.5–10 mm from the center.Citation82 NPR1 is a suppressor of cell death. NPR3 and NPR4, as BTB/POZ domain proteins, interact with both NPR1 and Cullin 3 E3 ligase and function as adaptor proteins to facilitate the poly-ubiquitination and subsequent degradation of NPR1 protein by the 26S proteasome.Citation83 NPR4 has a high affinity for SA while NPR3 has a low affinity for SA. High SA level at the center of cell death zone disrupts the interaction between NPR1 and NPR4, but favors the interaction between NPR1 and NPR3. NPR3 then promotes degradation of NPR1 to a level that is not sufficient to suppress cell death. In neighboring cells, the intermediate level of SA disrupts NPR1–NPR4 interaction, but is not high enough to promote NPR1–NPR3 interaction, so neither NPR3 nor NPR4 can mediate the degradation of NPR1. As a result, the high level of NPR1 in these neighboring cells suppresses the spreading of cell death and interacts with transcription factors to activate plant defense responses.Citation83
ROS and cell death
It is generally believed that the onset of HR is due to elevated level of reactive oxygen species (ROS). One major way of generating ROS during plant–pathogen interactions is through NADPH oxidases, which are called respiratory burst oxidase homologs (RBOHs) in plants. Cell death is greatly reduced in Arabidopsis atrbohD mutant and atrbohD/F double mutant plants compared with that in wild type plants following infection by an avirulent bacterial pathogen,Citation84 indicating that RBOHs and ROS are required for cell death induced by avirulent pathogens (). However, the relationship between ROS production and cell death is far more complicated than originally thought. For example, when infected with an avirulent oomycete pathogen, Arabidopsis atrbohD/F double mutant plants exhibited more cell death even though the ROS was eliminated.Citation84 When challenged by bacterial AvrRpm1, Arabidopsis nca1 (no catalase activity 1) and cat2 (catalase 2) mutants, which were supposed to have elevated ROS, surprisingly showed reduced cell death.Citation85 This result suggests the presence of additional pathways downstream of catalase other than through ROS in regulating cell death. The biological connection between ROS and PCD thus remains to be further studied.
Figure 2. Plant defense responses associated with ETI. Reactive oxygen species (ROS) production plays important roles in plant defense response during ETI (left). Plasma membrane-localized NADPH oxidases contribute to the production of ROS in plant apoplast, which triggers programmed cell death (PCD). In plants, these NADPH oxidase are called respiratory burst oxidase homologs (RBOHs). Catalase is an anti-oxidative enzyme that prevents accumulation of peroxisomal ROS. CAT2 (Catalase 2) and NCA1 (no catalase activity 1), which are required for catalase activities in plants, should in theory prevent PCD. However, both CAT2 and NCA1 contribute to autophagy-dependent PCD. In addition to the primary pro-death function of autophagy, autophagy downregulates ROS signaling in older plants or under long day condition. Autophagy limits PCD by a negative feedback pathway. Integrated model depicts selected transcriptional regulators associated with diverse R proteins to control the expression of defense genes (right). Association of R protein RRS1-R (WRKY52) with its cognate effector causes activation of defense genes, perhaps through derepression of transcriptional activity of WRKY domain or activation of other regulators. Upon avr pathogen infection, diverse activated R proteins MLA10, Pb1, SNC1, RPS4, and N are able to activate HvMYB6, OsWRKY45, TPR1, AtSPL6, and NbSPL6, respectively through protein-protein interactions, thus resulting in induction of defense genes. After perception of specific effector, MLA10 associates with HvWRKY1/2 repressors and thereby de-repress immune response. In addition, activated MLA10 enables to release HvMYB6 from HvWRKY1 repressor, which antagonistically associates with DNA binding domain of HvMYB6. The corepressor TRP1 represses transcription of two negative regulators of plant defense (DND1 and DND2), leading to induction of defense responses. Transcription of two tomato SIWRKY72 genes (SIWRKY72a and SIWRKY72b) are upregulated in defense response triggered by Mi-1 and these genes are required in Mi-1-mediated resistance. Both MED14 and MED16 are involved in induction of a large number of defense genes and resistance to RPT2. Blue lines indicate that diverse R proteins activate (arrows) or repress (truncated lines) activities of corresponding transcriptional regulators after perception of avr effectors. Dark red lines indicate that representative transcriptional (co)factors positively (arrows) or negatively (truncated lines) regulate transcription of downstream defense genes. Simple model for ELP2, SDG8 and MORC1 (CRT1)-mediated epigenetic control of transcriptome reprogramming in ETI is shown below. The histone acetyltransferase (HAT) activity of ELP2 for histone acetylation positively regulates defense genes including NPR1, PR1, PR2, PR5, EDS1, and PAD4. In addition, as a DNA demethylase, it reduces DNA methylation levels in NPR1 promoter and PAD4 coding regions after infection of Pst DC3000 avrRpt2, resulting in induction of NPR1 and PAD4. The histone lysine methytransferase SDG8, which trimethylated histone 3 lysine 36 (H3K36), is required for induction of LAZ5 (RPS4-like) and RPM1 R genes. Loss of SDG8 increases monomethylated H3K36 levels that probably is a general mark for transcription represson of a subset of R genes including RPM1. MORC1 (CRT1) causes heterochromatin condensation and is required for RPM1-mediated defense signaling. Avr Effectors, avirulence proteins; R, resistance proteins; PM, plasma membrane; RBOH, respiratory burst oxidase homolog; CAT2, catalase 2; NCA1, no catalase activity 1; ROS, reactive oxygen species; PCD, programmed cell death; RPM1, a CC-NB-LRR protein conferring resistance to Pseudomonas syringae pv Maculicola 1 (RPM1); RRS1-R, recessive resistance to Ralstonia solanacearum 1; MLA10, mildew A10 R protein; Pb1, Panicle blast 1; Mi-1, tomato cultivar Motelle; SNC1, a TIR-NB-LRR protein identified from suppressor of npr-1, constitutive 1 (SNC1); RPS4, resistance to P. syringae 4; N, Nicotiana TIR-NB-LRR receptor N; RPT2, resistance to P. syringae pv tomato 2; HvWRKY1/2, OsWRKY45 and SlWRKY72a/b, WRKY DNA-binding domain transcription factors in Hordeum vulgare (Hv), Oryza sativa (Os) and tomato Solanum lycopersicum (Sl); HvMYB6, R2R3 type MYB6-like transcription factor; MED14/16, mediator 14/16; NbSPL6 and AtSPL6, Squamosa promoter binding protein-like 6 (SPL6) transcription factors in Nicotiana benthamiana (Nb) and Arabidopsis thaliana (At); TPR1 (MOS10), Topless-related 1 (modifier of snc1, 10); DND1/2, Defense no Death1/2; ELP2, Elongator subunit 2; SDG8, SET (Su[var]3–9, E[z] and Trithorax conserved) DOMAIN GROUP 8; MORC1 (CRT1), Microrchidia 1 [MORC1] ATPase (compromised recognition of TCV-1 [CRT1]); Me, methylation; Ac, acetylation.
![Figure 2. Plant defense responses associated with ETI. Reactive oxygen species (ROS) production plays important roles in plant defense response during ETI (left). Plasma membrane-localized NADPH oxidases contribute to the production of ROS in plant apoplast, which triggers programmed cell death (PCD). In plants, these NADPH oxidase are called respiratory burst oxidase homologs (RBOHs). Catalase is an anti-oxidative enzyme that prevents accumulation of peroxisomal ROS. CAT2 (Catalase 2) and NCA1 (no catalase activity 1), which are required for catalase activities in plants, should in theory prevent PCD. However, both CAT2 and NCA1 contribute to autophagy-dependent PCD. In addition to the primary pro-death function of autophagy, autophagy downregulates ROS signaling in older plants or under long day condition. Autophagy limits PCD by a negative feedback pathway. Integrated model depicts selected transcriptional regulators associated with diverse R proteins to control the expression of defense genes (right). Association of R protein RRS1-R (WRKY52) with its cognate effector causes activation of defense genes, perhaps through derepression of transcriptional activity of WRKY domain or activation of other regulators. Upon avr pathogen infection, diverse activated R proteins MLA10, Pb1, SNC1, RPS4, and N are able to activate HvMYB6, OsWRKY45, TPR1, AtSPL6, and NbSPL6, respectively through protein-protein interactions, thus resulting in induction of defense genes. After perception of specific effector, MLA10 associates with HvWRKY1/2 repressors and thereby de-repress immune response. In addition, activated MLA10 enables to release HvMYB6 from HvWRKY1 repressor, which antagonistically associates with DNA binding domain of HvMYB6. The corepressor TRP1 represses transcription of two negative regulators of plant defense (DND1 and DND2), leading to induction of defense responses. Transcription of two tomato SIWRKY72 genes (SIWRKY72a and SIWRKY72b) are upregulated in defense response triggered by Mi-1 and these genes are required in Mi-1-mediated resistance. Both MED14 and MED16 are involved in induction of a large number of defense genes and resistance to RPT2. Blue lines indicate that diverse R proteins activate (arrows) or repress (truncated lines) activities of corresponding transcriptional regulators after perception of avr effectors. Dark red lines indicate that representative transcriptional (co)factors positively (arrows) or negatively (truncated lines) regulate transcription of downstream defense genes. Simple model for ELP2, SDG8 and MORC1 (CRT1)-mediated epigenetic control of transcriptome reprogramming in ETI is shown below. The histone acetyltransferase (HAT) activity of ELP2 for histone acetylation positively regulates defense genes including NPR1, PR1, PR2, PR5, EDS1, and PAD4. In addition, as a DNA demethylase, it reduces DNA methylation levels in NPR1 promoter and PAD4 coding regions after infection of Pst DC3000 avrRpt2, resulting in induction of NPR1 and PAD4. The histone lysine methytransferase SDG8, which trimethylated histone 3 lysine 36 (H3K36), is required for induction of LAZ5 (RPS4-like) and RPM1 R genes. Loss of SDG8 increases monomethylated H3K36 levels that probably is a general mark for transcription represson of a subset of R genes including RPM1. MORC1 (CRT1) causes heterochromatin condensation and is required for RPM1-mediated defense signaling. Avr Effectors, avirulence proteins; R, resistance proteins; PM, plasma membrane; RBOH, respiratory burst oxidase homolog; CAT2, catalase 2; NCA1, no catalase activity 1; ROS, reactive oxygen species; PCD, programmed cell death; RPM1, a CC-NB-LRR protein conferring resistance to Pseudomonas syringae pv Maculicola 1 (RPM1); RRS1-R, recessive resistance to Ralstonia solanacearum 1; MLA10, mildew A10 R protein; Pb1, Panicle blast 1; Mi-1, tomato cultivar Motelle; SNC1, a TIR-NB-LRR protein identified from suppressor of npr-1, constitutive 1 (SNC1); RPS4, resistance to P. syringae 4; N, Nicotiana TIR-NB-LRR receptor N; RPT2, resistance to P. syringae pv tomato 2; HvWRKY1/2, OsWRKY45 and SlWRKY72a/b, WRKY DNA-binding domain transcription factors in Hordeum vulgare (Hv), Oryza sativa (Os) and tomato Solanum lycopersicum (Sl); HvMYB6, R2R3 type MYB6-like transcription factor; MED14/16, mediator 14/16; NbSPL6 and AtSPL6, Squamosa promoter binding protein-like 6 (SPL6) transcription factors in Nicotiana benthamiana (Nb) and Arabidopsis thaliana (At); TPR1 (MOS10), Topless-related 1 (modifier of snc1, 10); DND1/2, Defense no Death1/2; ELP2, Elongator subunit 2; SDG8, SET (Su[var]3–9, E[z] and Trithorax conserved) DOMAIN GROUP 8; MORC1 (CRT1), Microrchidia 1 [MORC1] ATPase (compromised recognition of TCV-1 [CRT1]); Me, methylation; Ac, acetylation.](/cms/asset/5e6a59f0-5414-4e16-b577-25bd74d6df56/kvir_a_10929755_f0002.gif)
Autophagy and cell death
One important discovery related to ETI is the finding that PCD is regulated by autophagy (). Liu et al. showed that downregulation of autophagy genes ATG6 and ATG7 in Nicotiana benthamiana caused extended cell death when these plants were infected by TMV.Citation86 Later, the same group showed AvrRpm1-induced HR in ATG6 antisense plants also spread to uninfected tissue.Citation87 Both studies support a function for autophagy genes to restrict PCD to infection sites and negatively regulate PCD. However, a different study showed that ATG7 and ATG9 knockout plants exhibited suppressed initiation of TIR-NB-LRR type R proteins-mediated EDS1-dependent cell death, while CC-NB-LRR type R proteins-mediated NDR1-dependent cell death is autophagy-independent. Therefore, autophagic cell death regulated by ATG7 and ATG9 contributes to PCD mediated by certain type R proteins.Citation88 This dual role of autophagy in PCD is partially explained by studies from a third group, which showed that the positive or negative role of autophagy in PCD is dependent on the age of the plant leaves and photoperiod conditions used in the assays. Only older leaves of atg5 mutant plants showed spread RPM1-mediated cell death, which is believed to be a result of excessive ROS production and over-accumulation of SA.Citation89 In addition, the role of autophagy in PCD is dependent on the photoperiod conditions (), as atg2 mutant plants showed reduced cell death under short-day conditions, and exaggerated cell death when short-day-grown plants were moved to long-day conditions.Citation89 Furthermore, autophage-dependent cell death has been demonstrated to be regulated by ROS and catalaseCitation85 (). It will be of interest to the field of plant-microbe interactions to uncover the molecular mechanism underlying the regulatory role for autophage in PCD.
Transcriptional Reprogramming of Defense-Responsive Genes
WRKY transcription factors
ETI is a strong plant immune response associated with extensive transcriptional reprogramming that involves numerous transcriptional regulatorsCitation9,Citation10,Citation90,Citation91 (). Dynamic recruitment of transcriptional regulators into R protein-mediated signaling pathways has emerged as an important signaling event after the recognition of Avr effectors during ETI.Citation92,Citation93 Many WRKY transcription factors have been shown to play pivotal roles in regulating defense responses.Citation94,Citation95 TIR type R protein RRS1-R/WRKY52, which contains a C-terminal WRKY domain, interacts with the R. solanacearum type III effector PopP2 in the nucleus.Citation35,Citation68,Citation96,Citation97 RRS1-R/WRKY52 is identical to SENSITIVE TO LOW HUMIDITY 1 (SLH1), which, when inactive, functions as a transcription repressor of many downstream defense genes including PR1, PR2, PR5, EDS1, and PHYTOALEXIN-DEFICIENT4 (PAD4).Citation98 Upon recognition of PopP2, RRS1-R/WRKY52 induces resistance signaling, possibly by suppressing the negative function of its WRKY DNA-binding domain, or through activation of other positive transcription regulatorsCitation96 (). Future research on identification of the RRS1-R/WRKY52/SLH1 interacting components and target genes will facilitate better understanding of the regulatory role of RRS1-R in plant defense signaling.
In barley, HvWRKY1/2 TFs interact with R protein MLA10 upon activation by avirulence A10 effectorCitation69 (). Silencing and overexpression experiments have shown that these WRKY TFs act as transcriptional repressors of basal defense during compatible and incompatible interaction with pathogens, indicating that repression of these WRKY TFs-mediated negative regulation of MAMP-induced genes could be a way to trigger the MLA-mediated defense responses.Citation69 Likewise, the homologs of HvWRKY1/2 in Arabidopsis, known as AtWRKY18/40, have been identified in repressing basal defense responses to virulent hemibiotrophic P. syringae and virulent powdery mildew pathogen Golovinomyces orontii. AtWRKY18 and AtWRKY40 repress the expression of positive regulators of defense by direct binding to the W-box cis-elements in the promoters of these regulator genes such as EDS1, APETALA2/Ethylene Responsive Factor (AP2/ERF)-type TF gene RRTF1 and JASMONATE-ZIM-DOMAIN PROTEIN 8 (JAZ8).Citation69,Citation99 Indeed, MLA10 interacts with two antagonistically acting TFs, HvWRKY1 transcription repressor and HvMYB6 transcription activator. The activated MLA10 can release the positive regulator HvMYB6 from HvWRKY1 repression to initiate downstream transcriptional reprogramming during innate immune responsesCitation100 (). Additional defense genes directly modulated by WRKY1-MYB6 will need to be defined. These findings demonstrated that MLA10 activates defense gene transcription by interfering with WRKY repressors through direct interaction and releasing transcription activators from WRKY repression.
Differing from the majority of WRKY TFs, OsWRKY45 is a transcriptional activator and positively regulates rice resistance to panicle blast (). A CC-NB-LRR family R protein Pb1 stabilizes OsWRKY45 to avoid its degradation in a ubiquitin proteasome system.Citation101 It is possible that the stabilization of OsWRKY45 enhances its transcriptional activity and upregulates expression level of its encoding gene OsWRKY45, and also OsWRKY46 and RDX genes in blast resistance.Citation101,Citation102 In tomato, two SIWRKY72 type TFs were also shown to positively regulate basal resistance and resistance mediated by R protein Motelle (Mi-1)Citation103 ().
SPL6 transcription factor and TPR1 corepressor
Similar to the function of WRKY TFs in R protein Pb1 and Mi-1-mediated resistance, SQUAMOSA PROMOTER BINDING PROTEIN (SBP) domain-containing transcription factor SPL6 and corepressor TPR1 have been shown to play positive roles in TIR-NB-LRR R proteins, N and SNC1, -mediated plant defense responses, respectively (). SPL6 has been shown to directly bind to N protein and is required for N-mediated resistance to TMV. Arabidopsis SPL6 homolog, AtSPL6, activates expression of a subset of defense genes. It is required for TIR-NB-LRR type R protein RPS4-mediated resistance, but not required for the signaling of CC-NB-LRR type R proteins RPM1 and RPS2, suggesting that the regulatory role of SPL6 in ETI might be specific.Citation104 The transcription corepressor Topless-related1 (TPR1; previously identified as modifier of snc1, 10 [MOS10]), which interacts with SNC1 in vivo and in vitro, has been shown to be a positive regulator to activate SNC1-mediated resistance.Citation105 Two negative regulators of plant defense, Defense no Death 1 (DND1) and Defense no Death 2 (DND2), were among dozens of genes that are suppressed by TPR1 in defense responses.Citation105 Thus, the repression of negative regulators is implicated in SNC1-mediated downstream transcriptional reprogramming in ETI.
The Mediator complex
The Mediator (MED) complex is a conserved transcription regulator in eukaryotes, which acts as an adaptor between TFs and the RNA polymerase II (RNAPII) to regulate transcriptional reprogramming.Citation106 The core of mediator consists of more than 20 subunits, which are organized into three modules, namely head, middle and tail. To date, many characterized Mediator subunits (MED8, MED14, MED15, MED16, MED19a, MED21, and MED25) in Arabidopsis have been functionally characterized in plant immunity.Citation91,Citation107,Citation108 Two tail module subunits, MED14 and MED16, function as positive regulators in ETICitation91,Citation109 (). Both the med14 and med16 mutants were deficient in transcriptional reprogramming induced by avirulent bacterial pathogen P. syringae pv tomato (Pst) DC3000 (avrRpt2), and conferred susceptibility to both Pst DC3000 (avrRpt2) and Pst DC3000. Interestingly, the number of affected genes was smaller in med14 mutant plants than in med16 mutant post Pst DC3000 (avrRpt2) infection.Citation109 These data imply that MED14 and MED16 might regulate plant defense gene expression using different mechanisms during ETI.
Chromatin modifications
Rapid transcriptional changes in ETI can be extensively influenced by modulation of chromatin configuration utilizing different mechanisms, such as methylation of cytosine residues located in DNA, posttranslational modifications of histones and ATP-dependent chromatin remodeling. The Arabidopsis Elongator complex subunit2 (ELP2) was recently postulated as a genomic DNA demethylase, which is typically required for the demethylation and induction of NPR1 and PAD4 genes in response to avirulent DC3000 (avrRpt2) infectionCitation107 (). Also, ELP2 possesses histone acetyltransferase activity, which is essential for the expression of several defense genes (i.e., NPR1, PR2, PR5, EDS1, and PAD4)Citation107 (). In fact, ELP2 has been confirmed as a major positive regulator of ETI, which promotes RPS2 and RPS4-mediated resistance to pathogens and kinetically induces defense genes, likely through histone modification.Citation110 Being a histone methyltranserase, Arabidopsis ATX1, a Trithorax (TRX) family protein, mediates the methylation of lysine 4 in histone H3 protein through its SET (named after three Drosophila proteins SUPPRESSOR OF VARIEGATION 3–9 (SU[VAR] 3–9), ENHANCER OF ZESTE (E[Z]) and TRX-containing domain, and is responsible for the upregulation of defense genes (i.e., WRKY70, PR1, and R genes).Citation111 Infection assays showed that atx1 mutant was resistant to Pst DC3000 hrcC mutant strain rather than virulent DC3000, indicating that ATX1 is a positive regulator of basal resistance.Citation111 However, the potential role of ATX1 in ETI remains to be further determined, considering the altered expression of TIR-NB-LRR R genes and other defense-related genes in atx1 mutant. Remarkably, another histone methyltransferase, SET DOMAIN GROUP 8 (SDG8), which epigenetically upregulates expression of a TIR-NB-LRR (RPS4-like) gene in a gene-specific manner, was shown to be required for RPM1-mediated resistance in ArabidopsisCitation112 ().
A conserved tetratricopeptide repeat (TPR) domain-containing protein known as SRFR1, similar to transcription repressors in other organisms, negatively regulates multiple NB-LRR R proteins and might work to suppress target genes by directing the preacetylation of nucleosomes.Citation113 Another conserved transcription cofactor, AtTIP49a, was shown to interact with the N-terminal half of RPM1 (CC-NB-LRR) and RPP5 (TIR-NB-LRR), and may act as a negative regulator of two RPP (for Resistance to Peronospora parasitica) proteins-dependent disease resistance.Citation114 TIP49a was initially found to be associated with the TATA binding protein (TBP) complex. TBP complex negatively regulates the transcription of oncogene c-myc, which contributes to cancer genesis and is induced by hepatitis C-virus infection,Citation115 through modification of chromatin involved in the recruitment of histone deacetylases.Citation116 Thus, it will be interesting to study other examples of the recruitment of SRFR1 or TIP49a, as well as chromatin-remodeling complexes, to specific target genes in plants.
Chromatin superstructure
Mutation of Microrchidia (MORC) family ATPase genes, AtMORC1 and AtMORC6, results in the induction of DNA-methylated genes and transposable elements without affecting the DNA and histone methylation status. Based on data from Hi-C analysis, a method to study three-dimensional architecture of genomes,Citation117 the atmorc1 and atmorc6 mutants both showed decondensation of pericentromeric DNA regions.Citation118 AtMORC1 is the same gene as CRT1, which was found to be required for disease resistance to TCV mediated by HRT.Citation119 In addition, CRT1 physically interacts with several R proteins including RPS2, Rx, and RPP8, and also R protein chaperone HSP90.Citation120 Silencing of CRT1 homolog in N. benthamiana caused reduced cell death triggered by Pto and RPM1, and crt1 mutant plants showed compromised R gene-mediated defense responsesCitation120 (). Taken together, these studies suggest that regulation of chromosome superstructure plays an important role in proper function of ETI.
Future Perspectives
There is no doubt that many recent discoveries have helped the community to gain a much better understanding of ETI. Tremendous progress has been made in unraveling the molecular mechanisms of ETI, especially in the recognition of Avr effectors, activation of R proteins and transcriptional reprogramming of plant defense genes. New models, including guard hypothesis and decoy model, have been developed to explain the recognition mechanism of Avr effectors by R proteins, along with the discovery and characterization of new effectors and their corresponding R proteins. However, details of these recognition events at the molecular level remain to be further studied. The resolution of molecular structures of Avr and R proteins will increase our understanding of the interactions between Avr and R proteins, and their biological roles. For example, a structural study of the molecular basis for plant immunity activated by AvrPto confirmed the inhibition of Pto kinase activity by AvrPto.Citation121 This study challenged a widely held hypothesis that AvrPto activates Pto kinase activity and helped with a better understanding of Pto-mediated defense signaling. Structural studies of Avr and R proteins will also make it more accurate to predict potential interactions between Avr and R proteins. R proteins undergo conformational changes upon activation. Alteration of intramolecular interaction between its different domains results in the binding of ADP to inactive R proteins and the binding of ATP to active R proteins. The model for the activation of R protein by an ATP/ADP molecular switch is largely based on their similarities to animal nucleotide-binding oligomerisation domain (NOD) proteins.Citation122-Citation124 More data from structural studies of plant R proteins are needed to support the legitimacy of this model. Furthermore, structural studies of purified R protein complexes before and after pathogen infection could be the key to understanding the dynamics of R protein functions. Knowledge of the crystal structure of R proteins with their corresponding partners in their inactive and active status will greatly benefit the understanding of the ETI initiation.
Activation of R proteins leads to extensive transcriptional reprogramming of plant defense genes. Plant transcriptional regulators related to ETI comprise not only canonical DNA-binding TFs that act as transcription activators or repressors, but also cofactors including coactivators and corepressors that associate with DNA-binding and transcription regulatory components to fine-tune host transcription during ETI. Characterized transcription regulators have been shown to play crucial roles in manipulating transcriptional reprogramming of many defense-responsive genes through dynamic interaction with cognate R proteins and recruitment of factors to modify chromatin structure. However, novel transcription regulators and their specific regulatory roles in plant defense signaling remain to be discovered. Further decryption of interplay between different transcriptional and posttranscriptional modifications (e.g., methylation, acetylation, S-nitrosylation, phosphorylation, and ubiquitinylation) will shed light on the regulatory network of plant immunity. It is also noteworthy that extensive transcriptional changes during ETI may require alteration of chromatin configuration through orchestrating transcription associated factors, histone modification, and chromatin remodelers. Thus, to better understand the precise role of epigenetic regulation of R protein-mediated resistance, a study utilizing both classic genetic analysis and cutting-edge systemic approaches will be extremely helpful and necessary.
To better control plant disease in the field, one potential application of the knowledge we gained so far is to construct transgenic crop plants expressing exogenous R genes to reduce the risk of infection of the host plants by certain pathogens. However, overexpression of R genes in transgenic plants carries a tremendous fitness cost. One possible strategy to bypass this problem is to introduce R genes into crop plants under the control of pathogen inducible promoters. Furthermore, introducing an exogenous R gene could potentially expose plants to new diseases caused by nectrotrophic pathogens. Lorang et al. recently reported that victorin effector from necrotrophic pathogen Cochliobolus victoriae, which kills plants to obtain nutrients, binds to the target protein TRX-h5 to activate R protein LOV1-mediated cell death in order to cause diseases in Arabidopsis.Citation125 This provided an excellent example of a necrotrophic pathogen taking advantage of cell death caused by ETI. This area of research is a new emerging theme in plant-pathogen interaction research. Another study showed that effector ToxA, from necrotrophic fungal pathogen Stagonospora nodorum, promotes the susceptibility of host plants by inducing Tsn1-dependent cell death.Citation126 Tsn1 encodes a protein similar to R protein with a serine/threonine protein kinase (S/TPK) domain and NBS-LRR domains.Citation126 It is likely that more examples of this kind of interaction between a pathogen and host plant will be discovered soon. As discussed by Lorang et al., while scientists explore the possibility of utilizing R proteins to control plant diseases, it is also possible that the newly introduced R proteins will be targeted by effectors from necrotrophic pathogens, and benefit necrotrophic pathogen infections.Citation125
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Reference
- Hamann T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front Plant Sci 2012; 3:77; http://dx.doi.org/10.3389/fpls.2012.00077; PMID: 22629279
- Underwood W. The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci 2012; 3:85; http://dx.doi.org/10.3389/fpls.2012.00085; PMID: 22639669
- Agrios GN, Beckerman J. Agrios’ Plant Pathology, 6th ed. 2011.
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A 2007; 104:19613 - 8; http://dx.doi.org/10.1073/pnas.0705147104; PMID: 18042724
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008; 20:471 - 81; http://dx.doi.org/10.1105/tpc.107.056754; PMID: 18263776
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 2009; 60:379 - 406; http://dx.doi.org/10.1146/annurev.arplant.57.032905.105346; PMID: 19400727
- Alfano JR, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol 1997; 179:5655 - 62; PMID: 9294418
- Dangl JL, Dietrich RA, Richberg MH. Death Don’t Have No Mercy: Cell Death Programs in Plant-Microbe Interactions. Plant Cell 1996; 8:1793 - 807; http://dx.doi.org/10.1105/tpc.8.10.1793; PMID: 12239362
- Caplan J, Padmanabhan M, Dinesh-Kumar SP. Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 2008; 3:126 - 35; http://dx.doi.org/10.1016/j.chom.2008.02.010; PMID: 18329612
- Bhattacharjee S, Garner CM, Gassmann W. New clues in the nucleus: transcriptional reprogramming in effector-triggered immunity. Front Plant Sci 2013; 4:364; http://dx.doi.org/10.3389/fpls.2013.00364; PMID: 24062762
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol 2004; 42:185 - 209; http://dx.doi.org/10.1146/annurev.phyto.42.040803.140421; PMID: 15283665
- Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 2013; 64:839 - 63; http://dx.doi.org/10.1146/annurev-arplant-042811-105606; PMID: 23373699
- Takken FL, Albrecht M, Tameling WI. Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 2006; 9:383 - 90; http://dx.doi.org/10.1016/j.pbi.2006.05.009; PMID: 16713729
- Lukasik E, Takken FL. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 2009; 12:427 - 36; http://dx.doi.org/10.1016/j.pbi.2009.03.001; PMID: 19394891
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A 1998; 95:10306 - 11; http://dx.doi.org/10.1073/pnas.95.17.10306; PMID: 9707643
- Levy M, Edelbaum O, Sela I. Tobacco mosaic virus regulates the expression of its own resistance gene N. Plant Physiol 2004; 135:2392 - 7; http://dx.doi.org/10.1104/pp.104.044859; PMID: 15299123
- Radwan O, Mouzeyar S, Nicolas P, Bouzidi MF. Induction of a sunflower CC-NBS-LRR resistance gene analogue during incompatible interaction with Plasmopara halstedii. J Exp Bot 2005; 56:567 - 75; http://dx.doi.org/10.1093/jxb/eri030; PMID: 15545294
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci U S A 1998; 95:1663 - 8; http://dx.doi.org/10.1073/pnas.95.4.1663; PMID: 9465073
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 2005; 435:1122 - 5; http://dx.doi.org/10.1038/nature03630; PMID: 15973413
- Yang B, White FF. Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol Plant Microbe Interact 2004; 17:1192 - 200; http://dx.doi.org/10.1094/MPMI.2004.17.11.1192; PMID: 15553245
- Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 2013; 11:745 - 60; http://dx.doi.org/10.1038/nrmicro3120; PMID: 24129510
- Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 2007; 19:2929 - 39; http://dx.doi.org/10.1105/tpc.107.051821; PMID: 17890374
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012; 24:859 - 74; http://dx.doi.org/10.1105/tpc.111.095380; PMID: 22408077
- Boccara M, Sarazin A, Thiébeauld O, Jay F, Voinnet O, Navarro L, Colot V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog 2014; 10:e1003883; http://dx.doi.org/10.1371/journal.ppat.1003883; PMID: 24453975
- Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 2009; 60:139 - 64; http://dx.doi.org/10.1146/annurev.arplant.59.032607.092906; PMID: 19014346
- Holt BF 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science 2005; 309:929 - 32; http://dx.doi.org/10.1126/science.1109977; PMID: 15976272
- Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 2010; 6:e1001111; http://dx.doi.org/10.1371/journal.ppat.1001111; PMID: 20862316
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci U S A 2011; 108:14694 - 9; http://dx.doi.org/10.1073/pnas.1105685108; PMID: 21873230
- Gou M, Shi Z, Zhu Y, Bao Z, Wang G, Hua J. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J 2012; 69:411 - 20; http://dx.doi.org/10.1111/j.1365-313X.2011.04799.x; PMID: 21967323
- Genoud T, Buchala AJ, Chua NH, Métraux JP. Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis. Plant J 2002; 31:87 - 95; http://dx.doi.org/10.1046/j.1365-313X.2002.01338.x; PMID: 12100485
- Zeier J, Pink B, Mueller MJ, Berger S. Light conditions influence specific defence responses in incompatible plant-pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 2004; 219:673 - 83; http://dx.doi.org/10.1007/s00425-004-1272-z; PMID: 15098125
- Griebel T, Zeier J. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 2008; 147:790 - 801; http://dx.doi.org/10.1104/pp.108.119503; PMID: 18434604
- Wu L, Yang HQ. CRYPTOCHROME 1 is implicated in promoting R protein-mediated plant resistance to Pseudomonas syringae in Arabidopsis. Mol Plant 2010; 3:539 - 48; http://dx.doi.org/10.1093/mp/ssp107; PMID: 20053798
- Jeong RD, Chandra-Shekara AC, Barman SR, Navarre D, Klessig DF, Kachroo A, Kachroo P. Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc Natl Acad Sci U S A 2010; 107:13538 - 43; http://dx.doi.org/10.1073/pnas.1004529107; PMID: 20624951
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A 2003; 100:8024 - 9; http://dx.doi.org/10.1073/pnas.1230660100; PMID: 12788974
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 2000; 19:4004 - 14; http://dx.doi.org/10.1093/emboj/19.15.4004; PMID: 10921881
- Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 2000; 12:2019 - 32; http://dx.doi.org/10.1105/tpc.12.11.2019; PMID: 11090206
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013; 25:1463 - 81; http://dx.doi.org/10.1105/tpc.112.107201; PMID: 23548743
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A 2006; 103:8888 - 93; http://dx.doi.org/10.1073/pnas.0602577103; PMID: 16731621
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003; 112:379 - 89; http://dx.doi.org/10.1016/S0092-8674(03)00040-0; PMID: 12581527
- Mackey D, Holt BF 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002; 108:743 - 54; http://dx.doi.org/10.1016/S0092-8674(02)00661-X; PMID: 11955429
- Liu J, Elmore JM, Lin ZJ, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 2011; 9:137 - 46; http://dx.doi.org/10.1016/j.chom.2011.01.010; PMID: 21320696
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 2011; 9:125 - 36; http://dx.doi.org/10.1016/j.chom.2011.01.009; PMID: 21320695
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 2003; 112:369 - 77; http://dx.doi.org/10.1016/S0092-8674(03)00036-9; PMID: 12581526
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 2005; 121:749 - 59; http://dx.doi.org/10.1016/j.cell.2005.03.025; PMID: 15935761
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol 2003; 49:1537 - 46; http://dx.doi.org/10.1046/j.1365-2958.2003.03666.x; PMID: 12950919
- Liu J, Elmore JM, Coaker G. Investigating the functions of the RIN4 protein complex during plant innate immune responses. Plant Signal Behav 2009; 4:1107 - 10; http://dx.doi.org/10.4161/psb.4.12.9944; PMID: 20514222
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 2009; 7:e1000139; http://dx.doi.org/10.1371/journal.pbio.1000139; PMID: 19564897
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al, European Union Effectoromics Consortium. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011; 333:596 - 601; http://dx.doi.org/10.1126/science.1203659; PMID: 21798943
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 2012; 12:89 - 100; http://dx.doi.org/10.1038/nri3141; PMID: 22273771
- Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 2005; 8:383 - 9; http://dx.doi.org/10.1016/j.pbi.2005.05.010; PMID: 15939664
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 2011; 334:1401 - 4; http://dx.doi.org/10.1126/science.1211641; PMID: 22158818
- Bhattacharjee S, Halane MK, Kim SH, Gassmann W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 2011; 334:1405 - 8; http://dx.doi.org/10.1126/science.1211592; PMID: 22158819
- Lewis JD, Lee AH, Hassan JA, Wan J, Hurley B, Jhingree JR, Wang PW, Lo T, Youn JY, Guttman DS, et al. The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci U S A 2013; 110:18722 - 7; http://dx.doi.org/10.1073/pnas.1315520110; PMID: 24170858
- van der Hoorn RA, Kamoun S. From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell 2008; 20:2009 - 17; http://dx.doi.org/10.1105/tpc.108.060194; PMID: 18723576
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Molecular Basis of Gene-for-Gene Specificity in Bacterial Speck Disease of Tomato. Science 1996; 274:2063 - 5; http://dx.doi.org/10.1126/science.274.5295.2063; PMID: 8953034
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB. Initiation of Plant Disease Resistance by Physical Interaction of AvrPto and Pto Kinase. Science 1996; 274:2060 - 3; http://dx.doi.org/10.1126/science.274.5295.2060; PMID: 8953033
- Jia Y, Loh YT, Zhou J, Martin GB. Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato cultivars and encode active protein kinases. Plant Cell 1997; 9:61 - 73; http://dx.doi.org/10.1105/tpc.9.1.61; PMID: 9014365
- Frederick RD, Thilmony RL, Sessa G, Martin GB. Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol Cell 1998; 2:241 - 5; http://dx.doi.org/10.1016/S1097-2765(00)80134-3; PMID: 9734361
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 2006; 18:2792 - 806; http://dx.doi.org/10.1105/tpc.106.044016; PMID: 17028203
- Zhou JM, Chai J. Plant pathogenic bacterial type III effectors subdue host responses. Curr Opin Microbiol 2008; 11:179 - 85; http://dx.doi.org/10.1016/j.mib.2008.02.004; PMID: 18372208
- Zipfel C, Rathjen JP. Plant immunity: AvrPto targets the frontline. Curr Biol 2008; 18:R218 - 20; http://dx.doi.org/10.1016/j.cub.2008.01.016; PMID: 18334200
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 2008; 18:74 - 80; http://dx.doi.org/10.1016/j.cub.2007.12.020; PMID: 18158241
- Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 2007; 318:648 - 51; http://dx.doi.org/10.1126/science.1144956; PMID: 17962565
- Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 2007; 318:645 - 8; http://dx.doi.org/10.1126/science.1144958; PMID: 17962564
- Römer P, Strauss T, Hahn S, Scholze H, Morbitzer R, Grau J, Bonas U, Lahaye T. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol 2009; 150:1697 - 712; http://dx.doi.org/10.1104/pp.109.139931; PMID: 19448036
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 2010; 48:419 - 36; http://dx.doi.org/10.1146/annurev-phyto-080508-081936; PMID: 19400638
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A 2002; 99:2404 - 9; http://dx.doi.org/10.1073/pnas.032485099; PMID: 11842188
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007; 315:1098 - 103; http://dx.doi.org/10.1126/science.1136372; PMID: 17185563
- García AV, Blanvillain-Baufumé S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog 2010; 6:e1000970; http://dx.doi.org/10.1371/journal.ppat.1000970; PMID: 20617163
- Heidrich K, Tsuda K, Blanvillain-Baufumé S, Wirthmueller L, Bautor J, Parker JE. Arabidopsis TNL-WRKY domain receptor RRS1 contributes to temperature-conditioned RPS4 auto-immunity. Front Plant Sci 2013; 4:403; http://dx.doi.org/10.3389/fpls.2013.00403; PMID: 24146667
- Sohn KH, Zhang Y, Jones JD. The Pseudomonas syringae effector protein, AvrRPS4, requires in planta processing and the KRVY domain to function. Plant J 2009; 57:1079 - 91; http://dx.doi.org/10.1111/j.1365-313X.2008.03751.x; PMID: 19054367
- Shen QH, Schulze-Lefert P. Rumble in the nuclear jungle: compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J 2007; 26:4293 - 301; http://dx.doi.org/10.1038/sj.emboj.7601854; PMID: 17853890
- Deslandes L, Rivas S. The plant cell nucleus: a true arena for the fight between plants and pathogens. Plant Signal Behav 2011; 6:42 - 8; http://dx.doi.org/10.4161/psb.6.1.13978; PMID: 21258210
- Li Y, Tessaro MJ, Li X, Zhang Y. Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol 2010; 153:1425 - 34; http://dx.doi.org/10.1104/pp.110.156240; PMID: 20439546
- Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 2005; 17:1306 - 16; http://dx.doi.org/10.1105/tpc.104.029926; PMID: 15772285
- Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol 2005; 15:1129 - 35; http://dx.doi.org/10.1016/j.cub.2005.05.022; PMID: 15964279
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, García AV, Wirthmueller L, Després C, Parker JE, Zhang Y, et al. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 2009; 21:2503 - 16; http://dx.doi.org/10.1105/tpc.108.064519; PMID: 19700630
- van Doorn WG. Classes of programmed cell death in plants, compared to those in animals. J Exp Bot 2011; 62:4749 - 61; http://dx.doi.org/10.1093/jxb/err196; PMID: 21778180
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 2009; 23:2496 - 506; http://dx.doi.org/10.1101/gad.1825209; PMID: 19833761
- Muthamilarasan M, Prasad M. Plant innate immunity: an updated insight into defense mechanism. J Biosci 2013; 38:433 - 49; http://dx.doi.org/10.1007/s12038-013-9302-2; PMID: 23660678
- Enyedi AJ, Yalpani N, Silverman P, Raskin I. Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc Natl Acad Sci U S A 1992; 89:2480 - 4; http://dx.doi.org/10.1073/pnas.89.6.2480; PMID: 1549613
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012; 486:228 - 32; PMID: 22699612
- Torres MA, Dangl JL, Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A 2002; 99:517 - 22; http://dx.doi.org/10.1073/pnas.012452499; PMID: 11756663
- Hackenberg T, Juul T, Auzina A, Gwizdz S, Malolepszy A, Van Der Kelen K, Dam S, Bressendorff S, Lorentzen A, Roepstorff P, et al. Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 2013; 25:4616 - 26; http://dx.doi.org/10.1105/tpc.113.117192; PMID: 24285797
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005; 121:567 - 77; http://dx.doi.org/10.1016/j.cell.2005.03.007; PMID: 15907470
- Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 2008; 4:20 - 7; PMID: 17932459
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 2009; 137:773 - 83; http://dx.doi.org/10.1016/j.cell.2009.02.036; PMID: 19450522
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009; 21:2914 - 27; http://dx.doi.org/10.1105/tpc.109.068635; PMID: 19773385
- Elmore JM, Lin ZJ, Coaker G. Plant NB-LRR signaling: upstreams and downstreams. Curr Opin Plant Biol 2011; 14:365 - 71; http://dx.doi.org/10.1016/j.pbi.2011.03.011; PMID: 21459033
- An C, Mou Z. The function of the Mediator complex in plant immunity. Plant Signal Behav 2013; 8:e23182; http://dx.doi.org/10.4161/psb.23182; PMID: 23299323
- Rivas S. Nuclear dynamics during plant innate immunity. Plant Physiol 2012; 158:87 - 94; http://dx.doi.org/10.1104/pp.111.186163; PMID: 21951465
- Rivas S, Deslandes L. Nuclear components and dynamics during plant innate immunity. Front Plant Sci 2013; 4:481; http://dx.doi.org/10.3389/fpls.2013.00481; PMID: 24319451
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 2007; 10:366 - 71; http://dx.doi.org/10.1016/j.pbi.2007.04.020; PMID: 17644023
- Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol 2009; 150:1648 - 55; http://dx.doi.org/10.1104/pp.109.138990; PMID: 19420325
- Lahaye T. Illuminating the molecular basis of gene-for-gene resistance; Arabidopsis thaliana RRS1-R and its interaction with Ralstonia solanacearum popP2. Trends Plant Sci 2004; 9:1 - 4; http://dx.doi.org/10.1016/j.tplants.2003.11.002; PMID: 14729210
- Tasset C, Bernoux M, Jauneau A, Pouzet C, Brière C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog 2010; 6:e1001202; http://dx.doi.org/10.1371/journal.ppat.1001202; PMID: 21124938
- Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki K. A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J 2005; 43:873 - 88; http://dx.doi.org/10.1111/j.1365-313X.2005.02500.x; PMID: 16146526
- Pandey SP, Roccaro M, Schön M, Logemann E, Somssich IE. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J 2010; 64:912 - 23; http://dx.doi.org/10.1111/j.1365-313X.2010.04387.x; PMID: 21143673
- Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen QH. Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 2013; 25:1158 - 73; http://dx.doi.org/10.1105/tpc.113.109942; PMID: 23532068
- Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, Sugano S, Jiang CJ, Takatsuji H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc Natl Acad Sci U S A 2013; 110:9577 - 82; http://dx.doi.org/10.1073/pnas.1222155110; PMID: 23696671
- Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H. The nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J 2012; Forthcoming PMID: 23013464
- Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J 2010; 63:229 - 40; http://dx.doi.org/10.1111/j.1365-313X.2010.04232.x; PMID: 20409007
- Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, Dinesh-Kumar SP. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog 2013; 9:e1003235; http://dx.doi.org/10.1371/journal.ppat.1003235; PMID: 23516366
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A 2010; 107:13960 - 5; http://dx.doi.org/10.1073/pnas.1002828107; PMID: 20647385
- Mathur S, Vyas S, Kapoor S, Tyagi AK. The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol 2011; 157:1609 - 27; http://dx.doi.org/10.1104/pp.111.188300; PMID: 22021418
- Wang Y, An C, Zhang X, Yao J, Zhang Y, Sun Y, Yu F, Amador DM, Mou Z. The Arabidopsis elongator complex subunit2 epigenetically regulates plant immune responses. Plant Cell 2013; 25:762 - 76; http://dx.doi.org/10.1105/tpc.113.109116; PMID: 23435660
- Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD. A downy mildew effector attenuates salicylic Acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol 2013; 11:e1001732; http://dx.doi.org/10.1371/journal.pbio.1001732; PMID: 24339748
- Zhang X, Yao J, Zhang Y, Sun Y, Mou Z. The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J 2013; 75:484 - 97; http://dx.doi.org/10.1111/tpj.12216; PMID: 23607369
- DeFraia CT, Zhang X, Mou Z. Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J 2010; 64:511 - 23; http://dx.doi.org/10.1111/j.1365-313X.2010.04345.x; PMID: 20807211
- Alvarez-Venegas R, Abdallat AA, Guo M, Alfano JR, Avramova Z. Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2007; 2:106 - 13; http://dx.doi.org/10.4161/epi.2.2.4404; PMID: 17965588
- Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, Brodersen P, Hofius D, Petersen M, Mundy J. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog 2010; 6:e1001137; http://dx.doi.org/10.1371/journal.ppat.1001137; PMID: 20949080
- Kim SH, Kwon SI, Bhattacharjee S, Gassmann W. Regulation of defense gene expression by Arabidopsis SRFR1. Plant Signal Behav 2009; 4:149 - 50; http://dx.doi.org/10.4161/psb.4.2.7682; PMID: 19649196
- Holt BF 3rd, Boyes DC, Ellerström M, Siefers N, Wiig A, Kauffman S, Grant MR, Dangl JL. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev Cell 2002; 2:807 - 17; http://dx.doi.org/10.1016/S1534-5807(02)00174-0; PMID: 12062092
- Higgs MR, Lerat H, Pawlotsky JM. Hepatitis C virus-induced activation of β-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events. Oncogene 2013; 32:4683 - 93; http://dx.doi.org/10.1038/onc.2012.484; PMID: 23108410
- Hecht A, Kemler R. Curbing the nuclear activities of beta-catenin. Control over Wnt target gene expression. EMBO Rep 2000; 1:24 - 8; http://dx.doi.org/10.1093/embo-reports/kvd012; PMID: 11256619
- van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, Dekker J, Lander ES. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp 2010; http://dx.doi.org/10.3791/1869; PMID: 20461051
- Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science 2012; 336:1448 - 51; http://dx.doi.org/10.1126/science.1221472; PMID: 22555433
- Kang HG, Kuhl JC, Kachroo P, Klessig DF. CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe 2008; 3:48 - 57; http://dx.doi.org/10.1016/j.chom.2007.11.006; PMID: 18191794
- Kang HG, Oh CS, Sato M, Katagiri F, Glazebrook J, Takahashi H, Kachroo P, Martin GB, Klessig DF. Endosome-associated CRT1 functions early in resistance gene-mediated defense signaling in Arabidopsis and tobacco. Plant Cell 2010; 22:918 - 36; http://dx.doi.org/10.1105/tpc.109.071662; PMID: 20332379
- Xing W, Zou Y, Liu Q, Liu J, Luo X, Huang Q, Chen S, Zhu L, Bi R, Hao Q, et al. The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 2007; 449:243 - 7; http://dx.doi.org/10.1038/nature06109; PMID: 17694048
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 2004; 343:1 - 28; http://dx.doi.org/10.1016/j.jmb.2004.08.023; PMID: 15381417
- Albrecht M, Takken FL. Update on the domain architectures of NLRs and R proteins. Biochem Biophys Res Commun 2006; 339:459 - 62; http://dx.doi.org/10.1016/j.bbrc.2005.10.074; PMID: 16271351
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 2005; 434:926 - 33; http://dx.doi.org/10.1038/nature03465; PMID: 15829969
- Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng SC, Maier CS, Wolpert TJ. Tricking the guard: exploiting plant defense for disease susceptibility. Science 2012; 338:659 - 62; http://dx.doi.org/10.1126/science.1226743; PMID: 23087001
- Faris JD, Zhang Z, Lu H, Lu S, Reddy L, Cloutier S, Fellers JP, Meinhardt SW, Rasmussen JB, Xu SS, et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci U S A 2010; 107:13544 - 9; http://dx.doi.org/10.1073/pnas.1004090107; PMID: 20624958
