Abstract
Retrotransposons constitute a major part of the genome in a number of eukaryotes. Long-terminal repeat (LTR) retrotransposons are one type of the retrotransposons. Candida albicans have 34 distinct LTR-retrotransposon families. They respectively belong to the Ty1/copia and Ty3/gypsy groups which have been extensively studied in the model yeast Saccharomyces cerevisiae. LTR-retrotransposons carry two LTRs flanking a long internal protein-coding domain, open reading frames. LTR-retrotransposons use RNA as intermediate to synthesize double-stranded DNA copies. In this article, we describe the structure feature, retrotransposition mechanism and the influence on organism diversity of LTR retrotransposons in C. albicans. We also discuss the relationship between pathogenicity and LTR retrotransposons in C. albicans.
Introduction
As an opportunistic fungal pathogen of humans, Candida albicans usually cause fungal infections such as thrush, vaginitis, and life-threatening bloodborne candidiasis.Citation1 Of the 16 Mb haploid genome of C. albicans, 14.9 Mb has been assembled by Stanford University (Stanford DNA Sequencing and Technology Center), which facilitates understanding of the genetic structure, pathogenicity, adaptability, and evolution in C. albicans. Retrotransposons are mobile genetic elements capable of independent transposition through RNA intermediates.Citation1 Retrotransposons are widespread transposable elements in eukaryotes, and constitute a major part of eukaryotic genomes in many cases.Citation2-Citation7 For instance, retrotransposons constitute 42% of human genomesCitation8 and 75% of maize genomes.Citation9
Two primary subclasses of retrotransposons are present in eukaryotic cells: long-terminal repeat (LTR) retrotransposons and non LTR retrotransposons (LINEs, long interspersed nuclear elements, and SINEs, short interspersed nuclear elements).Citation10
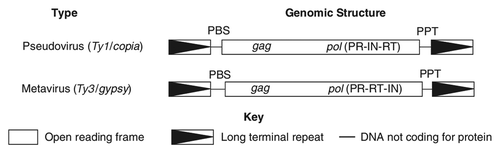
LTR retrotransposons in the yeast Saccharomyces cerevisiae have been extensively studied. According to the sequence similarity of reverse transcriptases (RTs)Citation11,Citation12 and the subunits of pol genes, there are two different types of LTR retrotransposons.Citation1 One is the Ty1/copia type, where the subunits are protease (PR), integrase (IN), reverse transcriptase (RT), and RNase H (RH) in order; the other is the Ty3/gypsy type, where the subunits are PR, RT, RH, and IN ().Citation13 In S. cerevisiae, six classes of retrotransposons, Ty1–5 and Ty3p, belong to 2 different types, respectively: Ty1/copia components (Ty1, Ty2, Ty4, and Ty5), and Ty3/gypsy components (Ty3 and Ty3p). In these retrotransposons families, Ty1, Ty2, Ty3, and Ty4 have transposition activity in S. cerevisiae as known.Citation14 This review summarizes the structure, mechanism, and influence on organism diversity of LTR-retrotransposons found in C. albicans.
LTR-Retrotransposon Families in C. albicans
Compared with the 6 families present in S. cerevisiae, 34 distinct LTR-retrotransposon families pertaining to the Ty1/copia and Ty3/gypsy groups have been found in C. albicans.Citation15 They are named by the Greek alphabet letters (α, β,… iota.), archaic Greek letters (sampi, san, etc.), and phonetically similar names of New Zealand birds (moa, tara, weka, etc.).Citation15 According to the internal regions, the first 16 LTRs have been defined as Tca1–Tca16 ().
Table 1. Properties of C. albicans retrotransposon LTR families
By comparing the LTR retrotransposons families in C. albicans and S. cerevisiae, we can find a great difference.Citation15 The different number of families illustrates that the two species experience disparity in retrotransposons element evolution.
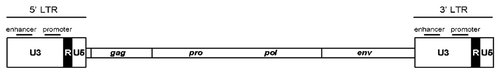
LTR retrotransposons contain two long-terminal repeats (LTRs), as shown in , at their ends, typically 250–600 bp in length, flanking a 5–7 kb long internal protein-coding domain.Citation16 Between the two LTRs are two open reading frames (ORFs): gag and pol. The gag ORF encodes the structural proteins that make up a virus like particle (VLP).Citation17 The pol ORF encodes the enzymes required for reverse transcription and integration. The enzymes are PR, IN, RT, and RH.
The LTRs of retrotransposons have the transcription termination signals and promoter, and are divided into three functional areas: U3, R, and U5.Citation17 U3 contains an enhancer and promoter region at the 3′ end of transcript; R contains both the start and termination sites for transcription;Citation18 U5 only exists in the transcript of 5′ terminal. The process of transcription is from the left LTR U3/R boundary to the right LTR R/U5 boundary to produce a RNA molecule with both ends of R region ().
Figure 2. LTRs can be subdivided into three regions: U3, R, and U5. U3 contains the enhancer and promoter sequences that drive viral transcription. R domain encodes the 5′ capping sequences (5′ cap) and the polyA (pA) signal.
Thirty-four LTR retrotransposon families in C. albicans share the following characteristics: (1) the length of the retrotransposon LTRs ranges from 127 to 780 bp with a mean length of 359 bp; (2) most elements have the terminal dinucleotides 5′-TG…CA-3′ and these dinucleotides tend to form part of larger terminal inverted repeats; (3) the total cope number of LTRs is 355; (4) both sides of more than half of the full-length LTR are short direct repeats representing target-site duplications (TSDs), most of which are 5 bp in length showing a tendency toward a purine in the first position and a pyrimidine in the last (data not shown);Citation15 and (5) these retrotransposon families in C. albicans generally vary widely in coding capacity as Ty families in S. cerevisiae and have a low copy number. Most families have the similar copy number to Ty5 (7 copies) LTR in S. cerevisiae, but considerably lower than that of Ty1/Ty2 (251 copies), Ty3 (41 copies), and Ty4 (32 copies).Citation15 Most LTR families in C. albicans only contain solo LTRs or LTR remnants in which the LTR-retrotransposon sequence has been retained and the intervening sequence has been lost. So far, just 3 intact retrotransposons have been founded. They are (1) Tca2, an active retrotransposon element which has a stop codon between two ORFs,Citation17 (2) Tca5, a Ty5 like retrotransposon,Citation19 and (3) Tca4, which belongs to Ty1/copia group and resembles to Tca2.Citation15 Several families of highly degenerate elements appear to be still capable of transposition, presumably via transactivation. Full-length retrotransposons are usually lost when two LTRs recombine with each other. This results in isolated solo LTRs at the original sites. A survey shows that 85% of Ty insertions in S. cerevisiae are solo LTRs or LTR fragments.Citation15 In C. albicans, for retrotransposon element, β, there are some 395 bp solo LTRs.Citation20 Both full-length retrotransposons and solo LTRs have short (4 or 5 bp) direct repeats on either end.Citation15 Most LTR retrotransposons generate 5 bp short direct repeats representing TSDs, as the Ty elements in S. cerevisiae.Citation15 For instance, 36% of the total S. cerevisiae Ty1–4 elements are flanked by TSDs.Citation21 Except for Ty3/gypsy elements (Ty3) that are flanked by 4 bp TSDs,Citation22 Ty5 elements have no TSDs. The condition of Ty5 suggests frequent recombination between these elements at the telomeres.Citation23 Recombination or mutation may have resulted in exchange of target site sequences between elements. A similar preference is apparent for the elements in C. albicans: 14 full-length LTRs are flanked by 4 bp direct repeats, like element pi,Citation10 weka has a confirmed 7 bp TSD. With analysis of the 5 bp TSDs in C. albicans and S. cerevisiae, base bias can be found: purine highly is enriched in the first site (55%); and pyrimidine is enriched in the last site (53%) and there is a strong bias for A and T: in the internal position 2 (72%), position 3 (76%), and position 4 (78%) (data not shown).Citation15
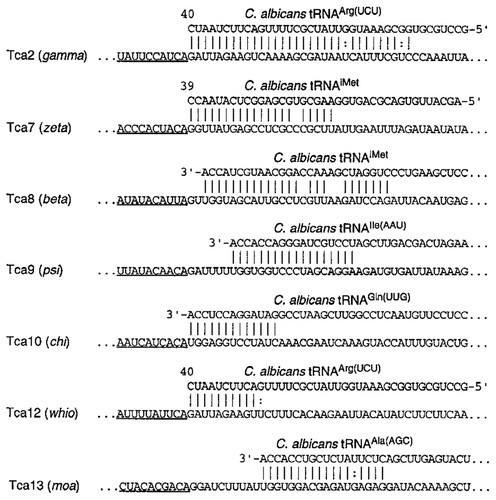
LTR retrotransposons have a potential tRNA primer-binding site. At the downstream from the left of 5′ LTR is a short polypurine sequence (8–49 nt), termed as the primer-binding sites (PBSs) (), which is a 10–20 nucleotide sequence that can base-pair with cytoplasmic tRNA molecule partly.Citation15 Fourteen different families of LTRs in C. albicans have connection with PBSs.
All PBSs share a common feature, the LTR terminus starting from 5′-TGG-3′ is base-pair with CCA sequence at the 3′ end of all tRNAs.Citation15 Goodwin et al.Citation15 had found the 3′ end of tRNA could be used as a primer to determine whether two LTRs were two independent insertions or a single retrotransposon. Furthermore, if the two LTRs belong to a family, they repeat in a direct orientation on a contig.Citation15 According to the above mentioned common features, 14 families of LTRs in C. albicans have been found sharing the relative PBSs. There are 7 different classes of these PBSs (): some have an extensive region which is homologous to an internal region of tRNAArg(UCU) (e.g., gamma), some have a short region which is homologous to the same tRNA (e.g., whio), some are homologous to a tRNAiMet internal region (e.g., zeta), and some are homologous to the 3′ ends of various tRNAs such as, Ala, Gln, Ile, and iMet tRNAs.Citation15 In some instances, the PBS is found to be located downstream of the LTR just 2–10 bp. A short gap between the LTR and the PBS is a common feature of gypsy-class retrotransposons.Citation24
Figure 3. The diversity of PBSs in C. albicans retrotransposons. The names of the retrotransposons and the associated LTRs are shown on the left. LTR sequences are underlined. The GenBank accession numbers of the tRNAs are as follows: tRNAArg, AF041470; tRNAiMet, AF069449; tRNAIle, Y08492; tRNAGln, AF180282; and tRNAAla, Y08493.
Mechanisms of Retrotransposition
A general phenomenon that resides in a number of eukaryotes is that adverse living conditions can activate retrotransposons. They can move from place to place in a genome by reverse transcription of a RNA transposition intermediate to enable the organism to adapt to the environment. It has been demonstrated that the retrotransposons generate new copies by reverse transcription of their RNA transcripts.Citation3
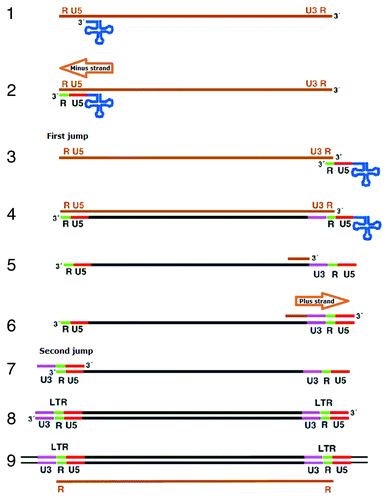
Similar to retroviruses in function, the LTRs of retrotransposons play a border role in complex reverse transcription procedure.Citation25 The two DNA strands are synthesized from opposite directions: tRNA binds to the site near the 3′ end of left LTR when synthesizing the first strand, while the polypurine track (PPT), a short purine-rich sequenceCitation17 immediately upstream of the right LTR, as a primer binds to the upstream of the right LTR when synthesizing the second strand.
Retrotransposons must synthesize mRNA at first, which can be translated into proteins related to replication, also acts as a replication template. At each end of the template, mRNA has a short repeat sequence (R, green) and U region ().Citation26 Primer tRNA can base-pair with PBS sequence. When tRNA anneals to PBS, minus strand DNA starts synthesizing under the catalysis of reverse transcriptase. DNA synthesized can reach the U5 region (red) at the 5′ end of the template and continue as the last few bases. The newly synthesized DNA can base-pair with the repeat sequence at the 3′ end of the genomic RNA.Citation26 Then, new strand DNA base-pair with 3′ end of the RNA template starting with U3 sequence, and continues until R and U5 at the 5′ end of RNA template. After that the RNase H degrades the RNA template leaving fragments at the poly-purine tract to prime second strand DNA synthesis. Using the new strand DNA as a template reverse transcriptase synthesizes another strand DNA until U5 and R. After that, the DNA fragment transfers to another end of new template. Synthesis then proceeds in both directions to give double-stranded DNA and LTR, making up of U5, R and U3, at each end.Citation26 Integrase inserts this into chromosomal DNA, and transcription initiating in one LTR and terminating in the other generates genomic RNA with terminal repeats.Citation26
Figure 4. Mechanisms of retrotranspositions. The RNA transposition intermediate (brown) of an LTR retrotransposon provide plus strand RNA. The RNA has a tRNA (blue) base-paired sequence, the PBS, near its 5′ end (1). Primer tRNA anneals to binding site on RNA (2). The first sequences to be copied are the unique sequence at the 5′ end of the RNA (U5, red) and a short repeat sequence (R, green) present at both ends of the RNA. In this step, single-stranded DNA R region pairs with 3′ terminus in the first jump (3). Reverse transcriptase starts synthesis minus strand DNA (4), starting with U3 (purple) adjacent to R, and continuing until U5 and R are copied a second time. tRNA primer is removed. The RNA template is degraded by RNase H leaving a fragment at the poly-purine tract to prime second strand DNA synthesis (5). In the second jump, reverse transcriptase transfers to the other end of minus strand (7). Synthesis then proceeds (8). Integrase inserts this into chromosomal DNA, and transcription initiating in one LTR and terminating in the other generates genomic RNA with terminal repeats (9).
Specific Description
Of the 34 distinct LTR-retrotransposon families in C. albicans, 16 families still retain some internal sequences which have very different coding capacity (Tca1–Tca16). Tca2, Tca4, and Tca5 are intact and possess all the characteristic features of functional retrotransposons. Tca3 and Tca8 have no full-length elements, but still retain long and continuous ORFs like other retroelements; Tca6 still contains ORF fragments in the internal region; Tca9 and Tca13 have 5 bp direct repeats at each end, intact PBSs and PPTs and identical LTRs; Tca10 is a compound element, whose LTR shares 99.5% identity and is flanked by a 5 bp direct repeat.Citation15 Some extensive ORFs, the PBS and PPT regions make up the ~2 kb long internal region.
The other 18 LTR families do not have the sequences similar to retrotransposons internal regions. These retrotransposon families may remain solo LTRs and LTR fragments, resulting in some of these LTRs escaping from detection.
Many researchers have studied a few LTR retrotransposons in C. albicans. Now we summarize the characteristics of Tca1, Tca2, Tca3, Tca4, and Tca5.
Tca1, an Inactive Element without ORF
Tca1, as the first C. albicans retroelement, is of 5614 bp and has 388 bp LTRs.Citation15 No significant ORF sequenceCitation27 presents in the internal region, suggesting that Tca1 is a degenerate and inactive element. It has been demonstrated that Tca1 has two loci in the strain SC5314. One is referred to as Tca1–1, and the other is designated as Tca1–2. The Tca1 elements of the two loci have more than 99% similarity in sequence and almost the same structure.Citation27Tca1–1 was isolated from a lambda phage genomic library clone called CJY-3.Citation28 This clone contains 3.25 kb and 4.8 kb long fragments (). Tca1–1 lacks any retrotransposon-like ORF. This differentiates Tca1–1 from Tca1–2. Tca1–2 was found in an approximately 6 kb clone, called CJY-4. It is highly similar to the insert of CJY-3, besides minor restriction site polymorphisms ().
Figure 5. Southern blot analysis of DNA from lambda clones CJY-3 and CJY-4. Genomic DNA from strain SC5314 or purified DNA from lambda clones CJY-3 and CJY-4 was digested with EcoRI and hybridized with the α-element probe.
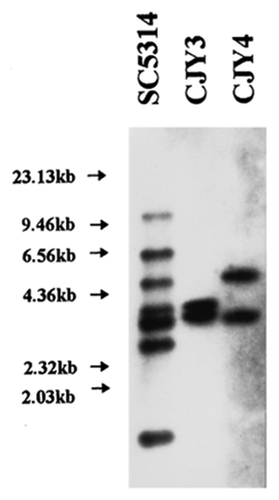
Figure 6. Restriction site maps of the genomic inserts from lambda clones CJY-3 and CJY-4 containing Tca1–1 and Tca1–2, respectively. The locations of elements (LTRs) are indicated by the boxed regions. Also indicated are the plus- and minus-strand primer binding sites (1PBS and 2PBS), the 5 bp direct repeats flanking the elements, the EcoRI fragments associated with each locus, and the 1.8 kb HindIII-EcoRI and 2.2kb EcoRV fragments that are used together as a hybridization probe of the internal region. The direction of transcription of Tca1 is from left to right.
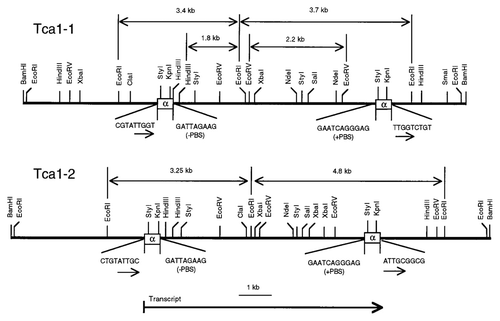
Nucleotide sequence analysis suggests that the insert of CJY-4 is flanked by identical direct repeat sequences, which was 99% identical to the LTRs of Tca1–1. Each LTR of Tca1–2 is flanked by 6 bp inverted repeat sequence (TGTTCG) like LTRs of other retrotransposons.Citation28 The sequence in the front of 5′ LTR and behind of 3′ LTR of Tca1–2 is ATTGC. This suggests that the sequence is a copy of integration target site.Citation29 The sequence is different from the one of Tca1–1, TTGGT.
Sequence analysisCitation30 of entire Tca1–2 has not found any extended ORFs or potential splice sites.Citation31 This suggests that Tca1–2 has been degenerated highly. Compared with about 1400 bp of insertion region of Tca1–1, Tca1–2 is just differing in less than 1% (data not shown).
Previous workCitation28 indicated that the transcription of Tca1–1 is affected by growth temperature. Northern blot analysis suggested that the transcription of Tca1 is also influenced by temperature in strains lacking Tca1–1 or Tca1–2.Citation27 Thus, Tca1 elements are temperature-regulated retrotransopsons. The expression level of Tca1 at 25 °C is higher than that at 37 °C.
It has been reported that the virulence determinants of a lot of pathogenic bacteria (Shigella app, Yersinia app, Vibrio cholerae, Bordetella pertussis, and Staphylococcus aureus) were regulated by temperature.Citation32 Antley et al.Citation33 found C. albicans cells grown at 25 °C are more virulent than those grown at 37 °C. As Ty elements can regulate adjacent gene transcriptionCitation34 the Tca1 may control adjacent genes temperature-dependently.Citation28 The expression of Tca1 could be similar to the invasion gene in Yersinia pseudotuberculosis.Citation35
Southern blot analysis suggests that deletion of Tca1 elements does not result in obvious growth defects and the recombination between LTRs occurs readily.Citation27 Furthermore, silencing a single element would lead to complete loss at that locus, suggesting that the Tca1 elements at both loci are hemizygous.Citation27 It is known that elements may be abundant in natural populations if they provide some advantage to the microorganism. However, there is no clear reason to maintain these degenerate elements. Neither copy of Tca1 choose silent chromatin as preferential integration region as the Ty5 retrotransposon in S. cerevisiae.Citation36-Citation39 Although nearly 40% of the strains lack Tca1 elements, solo LTRs in the genome suggest that they have existed in these strains for a time, and moreover, it is not essential to maintain Tca1 in natural populations.Citation28
Tca2 is an Abundant, Extrachromosomal DNA Molecule with Intact ORF
Tca2 is widespread in C. albicans and was first identified in the strain hOG1042, known as pCal. The pCal was a distinct band when the uncut C. albicans DNA was examined on an agarose gel, so it is an extrachromosomal retrotransposon.Citation17Tca2 belongs to Ty1/copia-type retrotransposon, and was originally identified as a linear double-stranded DNA molecule. It is the most abundant DNA copy (about 50 copies per cell).Citation16Tca2 is 6426 bp long and has 280 bp LTRs flanked by a six-nucleotide imperfect inverted repeat, TGTTGG…CCATCA. The internal domain usually has two long gag and pol-like ORFs.Citation16 ORF1 and ORF2 are separated by a stop-codon (UGA).Citation29 The structure feature is similar to mammalian retrovirusesCitation40 but is unique in LTR retrotransposons.Citation13 However, the pol gene can translate into protein. Forbes and coworkersCitation41 demonstrated that the LTR promoter is heat-shock induced. Like pol proteins, the expression of Tca2 pol protein is regulated by LTR promoter. There is a novel stop codon bypass mechanism to made gag-pol RNA translate pol proteins.Citation13 The mechanism is read-through suppression of the UAG codon.Citation17 In Tca2, the sequences between the gag and pol ORFs are an 8 bp purine-rich sequence, AAAACAGG, and a potential pseudoknot.Citation42 They are tightly followed the UGA stop codon, which is essential to suppression.
Tca2 has retrotranspositional activity, and is strongly dependent on the growth temperature.Citation16 There is substantially more extrachromosomal DNA at 37 °C than that at 27 °C. In addition, northern blot analysis on the transcriptional activity of Tca2 showed that the most prominent band is full-length transcripts. Interestingly, the number of full-length transcripts at 37 °C is more than that at 27 °C, suggesting that Tca2 has transposition activity and is in favor of higher temperature (37 °C). There is also a correlation between the level of Tca2 RNA and extrachromosomal DNA, suggesting that the synthesis of extrachromosomal DNA needs Tca2 RNA as intermediate. The level of Tca2 RNA is relative to the situation of Tca2 elements. For instance,Citation16 the levels of Tca2 RNA are different at various sites in genome. In addition, the levels of RNA are disparate in different strains.Citation16 The level of transcription is probably affected by strains, sequences and genomic context. Thus, we speculate that there may be close relationship between the Tca2 expression and virulence in the process of C. albicans evolution.
Tca3 has a Partial Coding Region
Tca3 is a Ty3/gypsy-like retrotransposon widely existing in C. albicans. It has 277bp LTRs and contains a little part of a coding region.
There are two major data sets of genome sequence have been determined for C. albicans strain SC5314.Citation43 The first is Assembly 6 and another is Assembly 19.
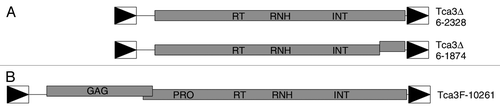
Assembly 6 is a high coverage sequence (~10-fold) in the genome. The database has two composites called as Tca3Δ,Citation44 sharing ~99% identity with each other. Both of them are ~4.55 kb, having an internal region flanked by LTRs (). The left LTR of Tca3Δ6–2328 has the same sequence as the right LTR of Tca3Δ6–1874, higher than its right LTR (99.4%); the right LTR of Tca3Δ6–2328 has 99.7% similarity to the left LTR of Tca3Δ6–1874 than its left LTR (99.0%).Citation44 also shows that in the internal regions of both Tca3Δ elements there are extensive ORFs. The internal region of Tca3Δ6–2328 contains one ORF almost whole length of itself, and in Tca3Δ6–1874 a frameshift ORF locate in the internal region. These ORFs contain the RT, RH, and IN domains like Ty3/gypsy retrotransposons in sequence.
Figure 7. Structures of Tca3 elements. (A) The two Tca3 elements in Assembly 6 of the Stanford C. albicans sequencing project database. (B) A full-length Tca3 element from C. albicans strain ATCC10261. In all panels shaded boxes represent the ORFs of the elements. The locations of the conserved domains are indicated. Offset boxes and vertical lines within the boxes represent frameshifts and premature stop codons, respectively. The LTRs are represented by the boxed triangles. The common scale is shown at the bottom.
Assembly 19 represents an initial draft of the diploid genome, sharing a similar Tca3 complement to Assembly 6.Citation44 However, it has some information which Assembly 6 does not contain, such as a composite on contig 19–10140.Citation44 Assembly 19 contains identical LTRs flanking a target site duplication instead of the internal region. Data suggest that the sequence of internal region is substituted by a run of Ns. Highly similarity to other Tca3 elements may lead the internal sequence co-assemble with them mistakenly.
Tca3 is roughly the same as other Ty3/gypsy elements in structure except for some special features. For example, elements of Tca3 seem to have a distinct mechanism for DNA synthesis which is involve in minus-strand DNA. Second, elements have a distant relationship with other Ty3/gypsy-like elements. Third, the ORFs of Tca3Δ elements have no obvious pol domain and shorter RT domain as typical Ty3/gypsy elements. Finally, losing the pol domain and large part of gag domain is another special feature.Citation44
A copy of Tca3 obtained from ATCC10261 that retains the gag/pol region is referred to as Tca3F-10261Citation44 (). It is found that Tca3Δ is more common than Tca3F. Tca3F is 6134 bp long. The internal region contains two long ORFs flanking by identical 313 bp LTRs. Tca3F-10261 has 99% identity with Assembly 6 Tca3Δ elements, except for the ~1.6 kb indel covering the gag/pol region, a 42 bp indel upwards to the large indel, and some variation in the 5′ untranslated regions.
Goodwin et al.Citation44 have found 5 of 6 strains emerged Tca3 bands, suggesting that Tca3 is quite common in C. albicans. In the five examined strains, only 1 to 3 bands were produced, which indicates that the elements are present in low copy number (data not shown). This might be the chance events in evolutionary process leading to excessive Tca3 elements produced. It may also be the result of selection, or may have connection with asexual reproduction in C. albicans.Citation45,Citation46 However, it is difficult to evaluate the relationship between different forms of Tca3 and host’s reproductive mode, on the base of indefinite reproduction in C. albicans.Citation47 Sequence comparison and Southern analysis have shown that Tca3 is much conserved in sequence and structure, either in loci or in strains, suggesting that the original deletion event has occurred not long ago.
Tca4, an Active Element, Contains Intact ORF
Tca4 is a Ty1/copia element closely related to Tca2,Citation16 which is flanked by 381 bp LTRs and contains an intact gag/pol ORFs. Tca4 is simple in sequence, suggesting that it has recently emerged. Like Tca2, Tca4 is a retrotranspositionally active element that strongly depends on the growth temperature. High temperature can induce Tca4 transcription.
Tca5, an Intact LTR Retrotransposon
Tca5 is of ~5.6 kb and 4218 bp of internal sequence flanked by identical 685 bp LTRs.Citation19 Only one single long ORF lies in the internal region. It is larger than most other yeast retrotransposon LTRs. Phylogenetic analysis and sequence comparison imply that Tca5 has close connection with Ty5 element in S. cerevisiae. The S. cerevisiae Ty LTRs range in length from 251 bp for Ty5Citation38 to 371bp for Ty4.Citation48,Citation49 The terminal inverted repeat sequences are the most conserved in Ty1-like elements, containing at least 5 nt: 5′-TGTTG…CAACA-3′. Therefore, the Tca5 retrotransposon begins with TG and ends with CA, as most retrotransposons do. Sequencing of the PCR product revealed the left LTR, which was identical to the right LTR, and these LTR are designated as omega.
Close behind the downstream of left LTR is PBS which is complementary to methionine tRNAiMet15 of initiator. Upstream of right LTR is a tract enriched in purines (ATGGGGAAG) and a similar sequence (ATGGGGAGG) at position 3132 bp.Citation19
The internal region contains a single and long uninterrupted ORF with a relatively low copy number, and this ORF extends from within the left LTR (382 bp) to within 60 bp of the right LTR, with no inframe stop codon or frame shift, predicting a protein with motifs characteristic of gag, protease, integrase, reverse transcriptase, and RNase H proteins of other retrotransposons.Citation19
However, it is unclear where the ORF might initiate translation in vivo.Citation19 There are several possible sites,Citation19 all of which are ATG in sequence. The first one within the ORF (382 bp from the 5′ end of the left LTR) is the unusually deep inset within the LTR. The second is also within the LTR three codons beyond the first. The third one within the ORF is unusually far beyond the end of the LTR, the three ATG are in-phase. Besides, there are several out-of-phase ATG triplets 5′ to this candidate.Citation19 Southern analysis (data not shown by original authors)Citation19 demonstrated that Tca5 is a low copy number retrotransposon, with very few solo LTRs. It is possible that these elements contribute to the heterogeneity of the Candida genome, either by recombination between the LTRs of one element or by chromosomal rearrangements between two elements. Tca5 may still be active which is supported by the detection of full-length transcript in northern blot.Citation19
In C. albicans, Tca5 is the only element showing significant similarity to Ty5.Citation19 It is a suitable candidate for comparisons because it has all the expected structures of an active element and is probably intact. Comparative genomic analysis between Tca5 and Ty5 elements will be helpful in understanding them.
LTR-Retrotransposons Improve Genetic Diversity
C. albicans is a diploid organism.Citation50 Clinical isolates of C. albicans show variform phenotypic characteristics associated with infection and virulence.Citation51-Citation57 These complex phenotypes suggest genetic diversity. However, C. albicans is an asexual fungus; it must have other mechanisms to generate various genomes. Retrotransposons have contributed to the evolution of genomes and genes in ways that go well beyond simply increasing genome size. It also affects genomes on a small scale, such as mobilization and integration at a locus directly, or recombination with heterotopic element indirectly. These changes contain gene inactivation, variance in transcription ability of gene, gene deletions and inversions.Citation29 It has been well documented that retrotransposons would mobilize in response to stress.Citation58-Citation60 Servant et al.Citation61 have shown that the relationship between transposable activity of Ty1 LTR retrotransposons and genome expression in S. cerevisiae. It seems to have a connection between stress reaction and the diversity of retrotransposons in C. albicans. Many organisms have the ability to recruit LTR-retrotransposons as alternative exons or promoters to drive genome evolution.Citation62-Citation65 The LTRs of Ty elements have the ability to modify genome transcriptionCitation61 and transposable elements can influence the expression of neighboring host genes.Citation61,Citation66 In gram-positive bacteria, circularized form of Tn retrotransposons can promote organisms resistance to antimicrobial.Citation67 There is an articleCitation33 about the relationship between virulence and reverse transposition of LTR retrotransposons in C. albicans. Take the Tca1 for example, the expression level of Tca1 is related to temperature, suggesting that Tca1 may place adjacent to the temperature-dependent genes. Since Tca1 exhibits higher expression at 25 °C than at 37 °C, Tca1 could upregulate the expression of genes required for the establishment of infection. Furthermore, in our original research, we found the transposition phenomenon of LTR-retrotransposon in strains which are resistant to miconazole (unpublished observation). It is suggested that there is a relationship between virulence and LTR-retrotransposons.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
L.Y. is supported by Natural Science Foundation of China (31000079). Y.C is supported by Natural Science Foundation of China (81173100) and Shanghai Basic Research Project (No11JC1415400). Y.J. is supported by China National 973 Program (2013CB531602) and Natural Science Foundation of China (81330083). The authors would like to apologize to all researchers whose important works were unable to be cited because of space limitations.
References
- Odds FC. Pathogenesis of Candida infections. J Am Acad Dermatol 1994; 31:S2 - 5; http://dx.doi.org/10.1016/S0190-9622(08)81257-1; PMID: 8077502
- Fradin C, Hube B. Tissue infection and site-specific gene expression in Candida albicans.. Adv Appl Microbiol 2003; 53:271 - 90; http://dx.doi.org/10.1016/S0065-2164(03)53008-8; PMID: 14696322
- Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell 1985; 40:491 - 500; http://dx.doi.org/10.1016/0092-8674(85)90197-7; PMID: 2982495
- Le QH, Wright S, Yu Z, Bureau T. Transposon diversity in Arabidopsis thaliana.. Proc Natl Acad Sci U S A 2000; 97:7376 - 81; http://dx.doi.org/10.1073/pnas.97.13.7376; PMID: 10861007
- Li WH, Gu Z, Wang H, Nekrutenko A. Evolutionary analyses of the human genome. Nature 2001; 409:847 - 9; http://dx.doi.org/10.1038/35057039; PMID: 11237007
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana.. Nature 2000; 408:796 - 815; http://dx.doi.org/10.1038/35048692; PMID: 11130711
- Flavell AJ, Pearce SR, Heslop-Harrison P, Kumar A. The evolution of Ty1-copia group retrotransposons in eukaryote genomes. Genetica 1997; 100:185 - 95; http://dx.doi.org/10.1023/A:1018385713293; PMID: 9440272
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al, International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001; 409:860 - 921; http://dx.doi.org/10.1038/35057062; PMID: 11237011
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet 1998; 20:43 - 5; http://dx.doi.org/10.1038/1695; PMID: 9731528
- Santana MF, Batista AD, Ribeiro LE, de Araújo EF, de Queiroz MV. Terminal repeat retrotransposons as DNA markers in fungi. J Basic Microbiol 2013; Forthcoming http://dx.doi.org/10.1002/jobm.201200453; PMID: 23440766
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 1990; 9:3353 - 62; PMID: 1698615
- Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 2008; 134:221 - 34; http://dx.doi.org/10.1016/j.virusres.2007.12.010; PMID: 18261821
- Neuvéglise C, Feldmann H, Bon E, Gaillardin C, Casaregola S. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res 2002; 12:930 - 43; http://dx.doi.org/10.1101/gr.219202; PMID: 12045146
- Carr M, Bensasson D, Bergman CM. Evolutionary genomics of transposable elements in Saccharomyces cerevisiae.. PLoS One 2012; 7:e50978; http://dx.doi.org/10.1371/journal.pone.0050978; PMID: 23226439
- Goodwin TJ, Poulter RT. Multiple LTR-retrotransposon families in the asexual yeast Candida albicans.. Genome Res 2000; 10:174 - 91; http://dx.doi.org/10.1101/gr.10.2.174; PMID: 10673276
- Holton NJ, Goodwin TJ, Butler MI, Poulter RT. An active retrotransposon in Candida albicans.. Nucleic Acids Res 2001; 29:4014 - 24; PMID: 11574684
- Matthews GD, Goodwin TJ, Butler MI, Berryman TA, Poulter RT. pCal, a highly unusual Ty1/copia retrotransposon from the pathogenic yeast Candida albicans.. J Bacteriol 1997; 179:7118 - 28; PMID: 9371461
- Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene 2005; 24:7656 - 72; http://dx.doi.org/10.1038/sj.onc.1209043; PMID: 16299527
- Plant EP, Goodwin TJ, Poulter RT. Tca5, a Ty5-like retrotransposon from Candida albicans.. Yeast 2000; 16:1509 - 18; http://dx.doi.org/10.1002/1097-0061(200012)16:16<1509::AID-YEA638>3.0.CO;2-R; PMID: 11113973
- Perreau VM, Santos MA, Tuite MF. Beta, a novel repetitive DNA element associated with tRNA genes in the pathogenic yeast Candida albicans.. Mol Microbiol 1997; 25:229 - 36; http://dx.doi.org/10.1046/j.1365-2958.1997.4521816.x; PMID: 9282735
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 1998; 8:464 - 78; PMID: 9582191
- Boeke JD, Stoye JP. Retrotransposons, Endogenous Retroviruses, and the Evolution of Retroelements. In: Coffin JM, Hughes SH, Varmus HE, eds. Retroviruses. Cold Spring Harbor (NY), 1997.
- Zou S, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 influences the organization of chromosome ends. Nucleic Acids Res 1996; 24:4825 - 31; http://dx.doi.org/10.1093/nar/24.23.4825; PMID: 8972872
- Chavanne F, Zhang DX, Liaud MF, Cerff R. Structure and evolution of Cyclops: a novel giant retrotransposon of the Ty3/Gypsy family highly amplified in pea and other legume species. Plant Mol Biol 1998; 37:363 - 75; http://dx.doi.org/10.1023/A:1005969626142; PMID: 9617807
- Boeke JD, Corces VG. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol 1989; 43:403 - 34; http://dx.doi.org/10.1146/annurev.mi.43.100189.002155; PMID: 2552899
- Finnegan DJ. Retrotransposons. Curr Biol 2012; 22:R432 - 7; http://dx.doi.org/10.1016/j.cub.2012.04.025; PMID: 22677280
- Chen Jy, Wang Q, Fu Z, Zhou S, Fonzi WA. Tca1, the retrotransposon-like element of Candida albicans, is a degenerate and inactive element. J Bacteriol 1998; 180:3657 - 62; PMID: 9658011
- Chen JY, Fonzi WA. A temperature-regulated, retrotransposon-like element from Candida albicans.. J Bacteriol 1992; 174:5624 - 32; PMID: 1324905
- Broach JR, Pringle JR, Jones EW. Molecular and cellular biology of the yeast Saccharomyces. 1991.
- Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res 1988; 16:1829 - 36; http://dx.doi.org/10.1093/nar/16.5.1829; PMID: 2832831
- Solovyev VV, Salamov AA, Lawrence CB. Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res 1994; 22:5156 - 63; http://dx.doi.org/10.1093/nar/22.24.5156; PMID: 7816600
- Maurelli AT. Temperature regulation of virulence genes in pathogenic bacteria: a general strategy for human pathogens?. Microb Pathog 1989; 7:1 - 10; http://dx.doi.org/10.1016/0882-4010(89)90106-X; PMID: 2682128
- Antley PP, Hazen KC. Role of yeast cell growth temperature on Candida albicans virulence in mice. Infect Immun 1988; 56:2884 - 90; PMID: 3049375
- Roeder GS, Fink G. Transposable elements in yeast. Mobile genetic elements Academic Press, Inc, New York 1983:299-328.
- Isberg RR, Swain A, Falkow S. Analysis of expression and thermoregulation of the Yersinia pseudotuberculosis inv gene with hybrid proteins. Infect Immun 1988; 56:2133 - 8; PMID: 2840402
- Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol 2001; 21:6606 - 14; http://dx.doi.org/10.1128/MCB.21.19.6606-6614.2001; PMID: 11533248
- Zhu Y, Zou S, Wright DA, Voytas DF. Tagging chromatin with retrotransposons: target specificity of the Saccharomyces Ty5 retrotransposon changes with the chromosomal localization of Sir3p and Sir4p. Genes Dev 1999; 13:2738 - 49; http://dx.doi.org/10.1101/gad.13.20.2738; PMID: 10541559
- Zou S, Ke N, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev 1996; 10:634 - 45; http://dx.doi.org/10.1101/gad.10.5.634; PMID: 8598292
- Zou S, Wright DA, Voytas DF. The Saccharomyces Ty5 retrotransposon family is associated with origins of DNA replication at the telomeres and the silent mating locus HMR. Proc Natl Acad Sci U S A 1995; 92:920 - 4; http://dx.doi.org/10.1073/pnas.92.3.920; PMID: 7846079
- Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci U S A 1985; 82:1618 - 22; http://dx.doi.org/10.1073/pnas.82.6.1618; PMID: 3885215
- Forbes EM, Nieduszynska SR, Brunton FK, Gibson J, Glover LA, Stansfield I. Control of gag-pol gene expression in the Candida albicans retrotransposon Tca2.. BMC Mol Biol 2007; 8:94; http://dx.doi.org/10.1186/1471-2199-8-94; PMID: 17961216
- Dam E, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry 1992; 31:11665 - 76; http://dx.doi.org/10.1021/bi00162a001; PMID: 1280160
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 1984; 198:179 - 82; http://dx.doi.org/10.1007/BF00328721; PMID: 6394964
- Goodwin TJ, Dalle Nogare DE, Butler MI, Poulter RT. Ty3/gypsy-like retrotransposons in Candida albicans and Candida dubliniensis: Tca3 and Tcd3.. Yeast 2003; 20:493 - 508; http://dx.doi.org/10.1002/yea.980; PMID: 12722183
- Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala FJ, Janbon F, Mallié M, Bastide JM. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc Natl Acad Sci U S A 1993; 90:9456 - 9; http://dx.doi.org/10.1073/pnas.90.20.9456; PMID: 8415722
- Lockhart SR, Fritch JJ, Meier AS, Schröppel K, Srikantha T, Galask R, Soll DR. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol 1995; 33:1501 - 9; PMID: 7650175
- Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell TG, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci U S A 1996; 93:12473 - 7; http://dx.doi.org/10.1073/pnas.93.22.12473; PMID: 8901606
- Janetzky B, Lehle L. Ty4, a new retrotransposon from Saccharomyces cerevisiae, flanked by tau-elements. J Biol Chem 1992; 267:19798 - 805; PMID: 1328182
- Stucka R, Schwarzlose C, Lochmüller H, Häcker U, Feldmann H. Molecular analysis of the yeast Ty4 element: homology with Ty1, copia, and plant retrotransposons. Gene 1992; 122:119 - 28; http://dx.doi.org/10.1016/0378-1119(92)90039-R; PMID: 1333437
- Olaiya AF, Sogin SJ. Ploidy determination of Canadida albicans.. J Bacteriol 1979; 140:1043 - 9; PMID: 391796
- Ghannoum M, Abu Elteen K. Correlative relationship between proteinase production, adherence and pathogenicity of various strains of Candida albicans.. J Med Vet Mycol 1986; 24:407 - 13; http://dx.doi.org/10.1080/02681218680000621; PMID: 3537262
- Mattia E, Carruba G, Angiolella L, Cassone A. Induction of germ tube formation by N-acetyl-D-glucosamine in Candida albicans: uptake of inducer and germinative response. J Bacteriol 1982; 152:555 - 62; PMID: 6752114
- McCourtie J, Douglas LJ. Relationship between cell surface composition, adherence, and virulence of Candida albicans.. Infect Immun 1984; 45:6 - 12; PMID: 6376361
- Mourad S, Friedman L. Pathogenicity of Candida.. J Bacteriol 1961; 81:550 - 6; PMID: 13773273
- Slutsky B, Buffo J, Soll DR. High-frequency switching of colony morphology in Candida albicans.. Science 1985; 230:666 - 9; http://dx.doi.org/10.1126/science.3901258; PMID: 3901258
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans.. J Bacteriol 1987; 169:189 - 97; PMID: 3539914
- Staib F. Proteolysis and pathogenicity of Candida albicans strains. Mycopathol Mycol Appl 1969; 37:345 - 8; http://dx.doi.org/10.1007/BF02129881; PMID: 5797623
- Anaya N, Roncero MI. Stress-induced rearrangement of Fusarium retrotransposon sequences. Mol Gen Genet 1996; 253:89 - 94; http://dx.doi.org/10.1007/s004380050300; PMID: 9003291
- Vasil’eva LA, Bubenshchikova EV, Ratner VA. [New evidence for the induction of mobile genetic element transpositions by severe heat shock]. Genetika 1998; 34:1243 - 50; PMID: 9879013
- Wessler SR. Turned on by stress. Plant retrotransposons. Curr Biol 1996; 6:959 - 61; http://dx.doi.org/10.1016/S0960-9822(02)00638-3; PMID: 8805314
- Servant G, Pennetier C, Lesage P. Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol Cell Biol 2008; 28:5543 - 54; http://dx.doi.org/10.1128/MCB.00416-08; PMID: 18591253
- Horie K, Saito ES, Keng VW, Ikeda R, Ishihara H, Takeda J. Retrotransposons influence the mouse transcriptome: implication for the divergence of genetic traits. Genetics 2007; 176:815 - 27; http://dx.doi.org/10.1534/genetics.107.071647; PMID: 17435252
- Landry JR, Rouhi A, Medstrand P, Mager DL. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol Biol Evol 2002; 19:1934 - 42; http://dx.doi.org/10.1093/oxfordjournals.molbev.a004017; PMID: 12411602
- Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem 2001; 276:1896 - 903; http://dx.doi.org/10.1074/jbc.M006557200; PMID: 11054415
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 2003; 19:530 - 6; http://dx.doi.org/10.1016/j.tig.2003.08.004; PMID: 14550626
- Knight SA, Labbé S, Kwon LF, Kosman DJ, Thiele DJ. A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev 1996; 10:1917 - 29; http://dx.doi.org/10.1101/gad.10.15.1917; PMID: 8756349
- Rice LB, Carias LL. Transfer of Tn5385, a composite, multiresistance chromosomal element from Enterococcus faecalis.. J Bacteriol 1998; 180:714 - 21; PMID: 9457879