Figures & data
Figure 1. A systems biology model for the brain-gut–microbiome interactions in mammals. The interconnected structural networks of the central nervous system influence via the autonomous nervous system to alter microbiota composition and function indirectly by regulating the microbial environment in the gut. The brain communicates with the gut microbiota indirectly through gut-derived molecules via afferent vagal and spinal nerve endings, or directly through microbe-generated signals. Alterations in these bidirectional interactions in response to perturbations like diet, medication, infections, and stress can alter the stability and behavior of this system resulting in brain-gut disorders.
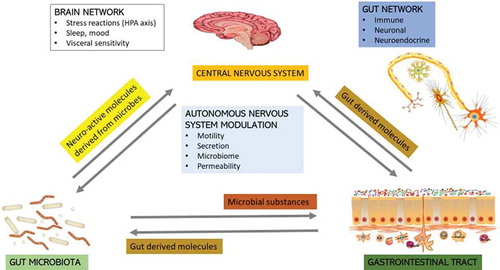
Figure 2. Factors affecting the gut microbiota in pigs. The diversity of gut microflora can be affected by a variety of factors. Sub-therapeutic doses of antibiotics used in commercial farming can negatively affect the microbiota. Similarly, different stressors such as high temperature, transportation, weaning, and overcrowding can also change the diversity of the microbiota for the worse. The inclusion of probiotics, prebiotics, and fiber appear to nullify these effects and improve diversity. Other factors, such as the age of the animal, its mode of the birth, breed and the environment it lives in, can influence the microbiota. Potentially pathogenic microbes are depicted in red and beneficial (and other commensals) microbes in green. The red arrows indicate a negative impact and a green arrow a positive impact. The gray arrows are an indication that different factors can affect the microbiota differently, either positively (e.g., if animals are bred in a healthy, growth-conducive environment) or negatively (e.g., following cesarean birth, devoid of any natural mother’s microbiota).
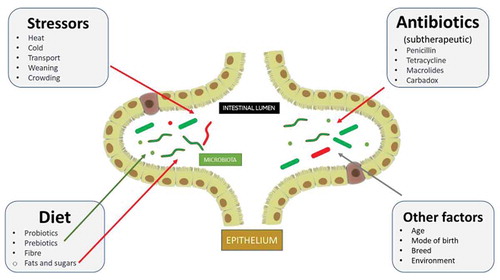
Table 1. Microbiome outcomes in studies examining pigs exposed to dietary and environmental modulations.
Figure 3. Inducible heat shock proteins. Dietary nutrients, gut microbiota components, and certain diseases can induce the formation of iHSPs (HSP25 and HSP70) in the GIT epithelium either directly from the microbiota or indirectly through the microbiota secretions and/or metabolites. This increases protection for the host against stressors like oxidation or inflammation in the epithelium.
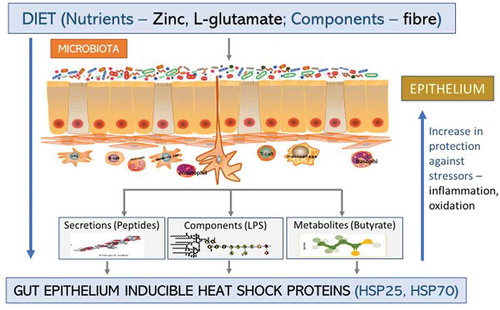
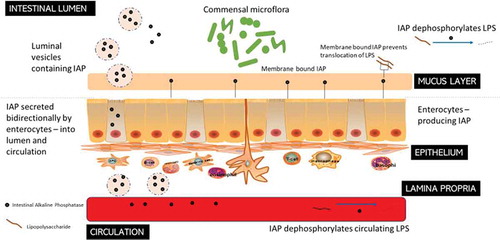
Figure 4. Intestinal Alkaline Phosphatase (IAP) – roles in the GIT. IAPs secreted by the enterocytes travel bi-directionally into the blood circulation and to the intestinal lumen where they act to dephosphorylate LPS from Gram-negative bacteria. Membrane-bound IAPs also prevent the translocation of the LPS through the mucus layer.
Data Availability Statement
No datasets were generated for this study.

