ABSTRACT
Pancreatic β-cells are connected to neighboring endocrine cells through the adherin proteins and gap junctions. Connexin 36 (Cx36) is one of the most well-studied and abundantly expressed gap-junction proteins within rodent islets, which is important in coordinated insulin secretion. The expression of connexins is regulated at various levels and by several mechanisms; one of which is via microRNAs. In past 2 decades, microRNAs (miRNAs) have emerged as key molecules in developmental, physiologic and pathological processes. However, very few studies have demonstrated miRNA-mediated regulation of connexins. Even though there are no reports yet on miRNAs and Cx36; we envisage that considering the important role of connexins and microRNAs in insulin secretion, there would be common pathways interlinking these biomolecules. Here, we discuss the current literature on connexins and miRNAs specifically with reference to islet function.
Increasing complexity in multi-cellular organisms requires a higher order of cell-cell coupling for optimal tissue function. In the islets of Langerhans, the secretion of insulin and other hormones needs to be carefully coordinated through membrane depolarization and calcium influx that drive the release of insulin in response to increasing concentrations of glucose. Gap junctions are cross-border gateways that connect adjoining cells, which readily transfer ions, metabolites and other small molecules between cells. Inter-cellular cross-talk is an essential component of normal cellular physiologic processes that integrate a range of environmental cues from their interactions with soluble factors, signaling molecules, extracellular matrix components as well as from their neighboring cells. A defect in signaling across such gap junctional proteins is implicated in various diseases, including diabetes. The goal of this article is to discuss the existing literature on connexin expression and islet cell function, then to introduce the literature on regulation of non-islet connexin expression by microRNAs and finally to emphasize the need for future research in regulation of islet-enriched connexins by microRNAs. Even though connexins in the pancreas have not been shown to be regulated by pancreas-specific/-enriched microRNAs, there is plenty of evidence suggesting the role of each of them (connexins and microRNAs) in insulin secretion, an important function of pancreatic islet cells. Bioinformatically, one can predict different microRNAs targeting pancreatic connexins and therefore it is very likely that connexins and microRNAs could be interlinked with each other in regulating pancreatic insulin secretion. We hope that future studies, which investigate microRNA-mediated-regulation of connexins in islet cells and their role in β-cell dysfunction and diabetes would lead to developing novel therapies for improving islet cell function, and survival, through regulation of gap junctional coupling.
Gap junctions and connexins
A pancreatic β-cell as well as other specialized endocrine cells, represent a highly active entity where several molecules are synthesized and degraded continuously. Most of the cellular physiologic processes are very well regulated and this regulation occurs at multiple levels. In a multicellular organism, where several cell types give rise to a single tissue that then serves a particular function for survival of that organism, it is extremely important that all cells efficiently communicate with each other. This is also true for maximizing the functional capacity of mini organs such as the islets of Langerhans. Most cells communicate with each other via secretion of effector molecules such as proteins, peptides, nucleic acids, hormones and ligands, and by responding to them. This communication can be either contact-independent (paracrine, endocrine and neuronal) or contact-dependent. In a contact-dependent communication, two of the cells need to be in a close vicinity to one another and should be connected to each other via a ligand-receptor complex or via gap junctions or via other cell adhesion molecules.
Gap junctions are important for the cell to cell communication, intercellular signaling and coordinated cellular functions.Citation1,3 In eukaryotes, metabolites, nutrients, signaling molecules; eg. small RNAs,Citation4 ions as well as electrical signals are exchanged via gap junctions, thus maintaining cellular homeostasis.Citation5 Apart from their role in cellular function, gap junctions are also important for normal development, proliferation, and differentiation of stem/progenitor cells. In vertebrates, connexins are building blocks of gap junctions, while innexins are proteins that form gap junctions in invertebrates.Citation6 There are 21 connexin proteins in humans and 20 connexins in mice.Citation7 Another family of connexin-like proteins; the pannexins, is known to be essential in mammals and consists of 3 members.Citation8
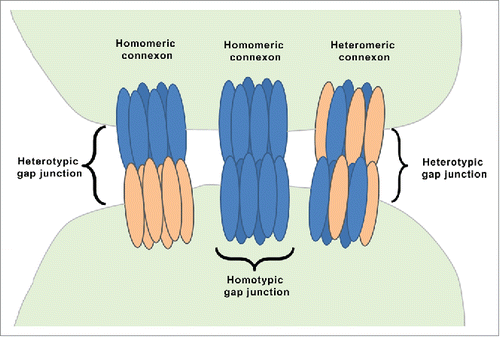
A single connexin protein is a non-glycosylated, transmembrane molecule that spans the plasma membrane 4 times and has both C- and N-terminals in the cytoplasm. Six connexin protein units bunch together in a cylindrical fashion and form a single connexon/hemichannel. Two hemichannels, one contributed by each of the adjoining cells, connect with each other and create a hydrophilic gap junction that allows transfer of small molecules (up to 900 daltons). Accordingly, the gap junctions are called as homotypic gap junction and heterotypic gap junction (). In homotypic gap junction, both hemichannels/connexons are identical, while heterotypic gap junction consists of 2 different connexons.Citation9 There are 2 types of connexons; homomeric and heteromeric connexon (). Homomeric connexons have identical connexins while heteromeric connexons are composed of different types of connexins.Citation9 It is also observed in some cases that a hemichannel does not connect to another cell but could open in the extracellular environment, facilitating the transports of ions, ATP and glutamate.Citation1,10
Figure 1. Organization of gap junction proteins. This is a schematic presentation of different types of connexons. A homomeric connexon refers to a connexon formed when 6 identical connexin proteins form the pore for a gap junction. A heteromeric connexon refers to a connexon formed between different connexin subunits. Two identical connexons form a homotypic gap junction, whereas a heterotypic gap junction consists of 2 different hemichannels.
Cell to cell communication via gap junctions is seen in almost all cells excluding erythrocytes, circulating lymphocytes and skeletal muscle cells.Citation2 Any changes or inhibition of gap junction-mediated cellular cross-talk leads to abnormal biology potentially contributing to the development of diseases including cancer and diabetes. Connexins have been well known as tumor supressors in cancer biology, as reviewed elsewhere.Citation11 With reference to the islet-enriched connexin; i.e. Cx36, it is well known that the gene encoding Cx36 is located on the 14q region of chromosome 15, which is a susceptibility locus for Type 2 diabetes, indicating a possible role in reduced β-cell function associated with progression to Type 2 diabetes.Citation12,13 Thus, there is significant evidence to understand the role of connexin dysfunction in disease. Although causality remains to be conclusively identified, the current evidence recognizes the need for normal connexin expression in optimal tissue function. Connexin proteins have a very short half-life of around 3 hours and are internalized or degraded by lysosomes or proteasomes.Citation1,2 Connexin expression and gap junctional intercellular communication are regulated at many levels including electrochemical gradient across the chanel, change in pH, change in the concentration of calcium ion, phosphorylation of connexin molecules, rearrangement of the structure as well as voltage and chemical gating.Citation2,3
Gap junctions in the pancreas
The pancreas is a unique organ wherein its endocrine portion; the islets of Langerhans, is embedded within the exocrine counterpart (acini and ducts). Islets consist of 5 different endocrine cell types; α, β, δ, pp and ϵ cells. The most abundant islet cell type is the β-cell, which produce and secrete insulin – the islet hormone that works toward reducing circulating glucose concentrations. Although there are significant differences observed between human and rodent islet architecture,Citation14-16 β-cells are always in contact with other β-cells or other endocrine pancreatic (non-β) cells and “hard-wired” or connected via specific connexins. Synchronized and pulsatile secretion of insulin requires co-ordination and electrical coupling with adjacent cells for transfer of the depolarizing signal from the adjacent endocrine cells in the islet to achieve normal insulin secretion; a process that is known to be coordinated via gap junction signaling.Citation17 Connexin 43 (Cx43) was demonstrated to be important in rat pancreas function;Citation18 however later on it was demonstrated that Cx36 is the most abundant and exclusive connexin expressed in pancreatic insulin-producing cellsCitation19-21 while Cx43 and Cx45 are expressed by vascular endothelial cells that are abundant in islets.Citation22,23 Recently, connexin 30.2 has been demonstrated to be present in mouse β-cells along with Cx36,Citation24 however, its functional role in insulin secretion is not yet confirmed. Connexins 26 and 32 have been reported in pancreatic exocrine/acinar cells during mouse pancreas development.Citation19 Cx36 is shown to be β-cell specific and is seen between adjacent β-cells,Citation21 while a probability of heterotypic gap junction consisting of Cx36 and Cx43 is suggested that may mediate β-cell and intra-islet endothelial cell interactions.Citation25
Cx36 knockout mouse islets fail to demonstrate intracellular calcium oscillations as well as the synchronous and pulsatile release of insulin.Citation26-28 Similar observations were reported in an in vitro manipulated (MIN6) cell line.Citation29 Cx36 knockout mice have normal fasting glucose, but display abnormal glucose clearance following an intraperitoneal glucose tolerance test, suggesting glucose intolerance.Citation17 There are also contradictory reports on the increase in basal insulin release in Cx36 deficient islets.Citation27,30 Studies in Cx36 knockout animals have indicated that these gap junctions in β-cells not only regulate insulin secretion but also regulate intra-islet blood flow.Citation31 High fat fed mice, which show insulin resistance, obesity and pre-diabetes have a significant reduction in islet Cx36 protein and around 30% less β-cell to β-cell coupling.Citation32 These data suggest that Cx36 gap junctions are affected and may contribute to islet β-cell dysfunction during the progression from impaired glucose tolerance to Type 2 diabetes. Although the role of connexins in β-cell dysfunction is well established, their role in β-cell survival is not demonstrated as yet. It is well understood that β-cell dysfunction precedes their death in progression to Type 2 diabetes.Citation33 The role of connexins in β-cell survival would need long-term assessment of β-cells from connexin-specific knockout mice during different stages of progression to Type 2 diabetes and insulin-requiring Type 2 diabetes. Overall, there is significant evidence to confirm the role of Cx36-dependent intercellular communication in glucose-stimulated insulin secretion (GSIS) and in β-cell dysfunction, leading to the development of Type 2 diabetes.
As mentioned above, connexin expression is regulated at multiple levels. However, there is not much information on regulation of Cx36 in islets. In the retinal AII amacrine cells, Cx36 is phosphorylated by protein kinase A (PKA) and it results in reduced coupling, decreased permeability across the Cx36 gap junction in these cells mediating visual adaptation.Citation34 Recently, another protein kinase PKCδ is shown to alter Cx36 coupling in islet cells and is believed to be a mechanism of islet dysfunction during cytokine exposure leading to diabetes.Citation35 Another study where islets were co-cultured with endothelial progenitor cells in vitro, reported decreased expression of Cx36 with increase in basal insulin secretion.Citation36 Even though the exact molecular mechanism was not investigated, a possibility of crosstalk between β-cells and endothelial cells is evident. All these reports point to the intricate regulatory processes within islet cells including those controlling connexin expression and coupling in pancreatic islets. Recently, microRNAs have been shown to regulate connexin expression and function in multiple non-islet tissues and in several species;Citation37-40 however, no reports on microRNA regulation of islet-specific/-enriched connexins are available till date. Considering the existence of such a mechanism in non-islet cell systems, this article presents a review of the known microRNAs that target connexins and attempts to initiate discussion in this area of biology that we think would emerge to be an important component of islet cell organization and function.
microRNAs
microRNAs are small, non-(protein) coding RNA molecules that are about 22 nucleotides in length and act as negative regulators of gene expression.Citation41 They are synthesized as a long transcript (primary miRNA) in the nucleus, which undergoes 2 rounds of processing by enzyme complexes (Drosha and Dicer in nucleus and cytoplasm respectively) to generate mature miRNAs in the cytoplasm.Citation42,43 These mature miRNAs are single-stranded molecules, which act at post-transcriptional level via incorporation into RISC (RNA-induced silencing complex) and regulate the target mRNA expression largely through 2 processes; i) degradation of target mRNA or ii) translational inhibition. Most miRNAs target 3′UTR of the mRNAs and in some cases its 5′UTR or the coding region, functioning as key regulators of developmental, physiologic and pathological conditions.Citation45,46
It is now known that microRNAs themselves are regulated by various other mechanisms including RNA-binding proteins, SNPs, methylation, miRNA-editing and circadian rhythms as reviewed elsewhere.Citation47 It has been demonstrated that a RNA-binding protein Deadend-1 (Dnd1) can physically block the access to a miRNA target site, thereby sterically hindering the normal function of RISC.Citation48 Although not much is known about the structural aspects of miR-RISC target recognition and Dnd1 binding, such molecules may offer another layer of regulation. It is also speculated that Dnd1 may change the subcellular localization of mRNA molecules, taking it out of the reach of its targeting miRNAs. Indeed, Dnd1 has been shown to localize to discrete perinuclear granules in primordial germ cells.Citation49
miRNAs are required for normal pancreas development, regeneration and function.Citation41,Citation50-52 miR-7 and -375 are some of the most abundant miRs within the pancreasCitation53 and are shown to regulate α and β-cell mass,Citation54,55 insulin expression,Citation56 insulin secretion,Citation57,58 as well as β-cell secretome.Citation59 Apart from these, several other miRNAs are implicated in regulating pancreas development by targeting important transcription factors.Citation41,50 Several microRNAs have been shown to be altered during β-cell dysfunction and/or apoptosis; especially miR-34a, miR-21, miR-29, and miR-146 have been reported to date;Citation51,Citation60-63 as well as in diabetes progression/complications.Citation64-66 Insulin secretion is also regulated by several different miRNAs, apart from the miR-7 and miR-375 mentioned above. These include miR-30d that targets MAP4K4,Citation67 miR-124a targeting sirt1, NeuroD1, FoxA2 and Rab27a,Citation68 miR-96 that targets Noc2 and granuphilin,Citation69 miR-33a/b targeting ABCA1Citation70 and multiple others. Although these studies underscore the importance of miRNAs in pancreas development, function, and disease (diabetes) progression, there are no reports yet on microRNA regulation of connexins in the pancreatic β-cells.
microRNAs and connexins
Till date, there are few studies demonstrating microRNA-mediated regulation of connexins. Most of these demonstrate regulation of Cx43, one of the most commonly expressed connexins in the body.Citation71,72 Bioinformatics analysis of microRNA and connexin interactions also predict Cx43 to be extensively regulated by miRNAs compared with other gap junction proteins.Citation73
In one of the reports, miR-206 and miR-1 microRNAs are shown to negatively regulate Cx43 during in vitro myoblast fusionCitation37 and a similar mechanism is implicated during muscle development in vivo. Elevated expression of miR-1, a muscle-specific miRNA, is observed during coronary artery disease and arrhythmia.Citation74,75 The proposed mechanism involves post-transcriptional repression of Kir2.1 and Cx43, thereby lowering conduction potential and depolarization of cardiac muscles. Three miRs (miR-1, -206 and -133) are also seen to be upregulated during in vitro myoblast differentiation with a concomitant reduction in Cx43 mRNA, suggesting that Cx43 is a potential target of these microRNAs.Citation76 MiR-206 is observed to be involved in osteoblast differentiation, where its abundance decreases during normal differentiation.Citation77 Overexpression and knockdown studies of this miRNA indicated that Cx43 is a target of miR-206 during bone formation. MiR-1 regulates Cx43 in cardiac hypertrophyCitation78 and the interaction between these 2 molcules is also important in bladder cells, thereby controlling bladder development and sensitivity.Citation79 Cx43 is also reported to be targeted by miR-218 in cancer cell lines,Citation80 miR-145 in corneal epithelial progenitor cells,Citation81 miR-221/222 cluster in glioblastoma cells,Citation38 miR-20a in prostate cancer cell linesCitation82 and miR-19a/b in murine cardiac cells.Citation83 Among other connexins, Cx40 is shown to be regulated by miR-208a, which is also necessary for normal cardiac conduction and function.Citation84,85
miRNAs transfer via connexins/gap junction
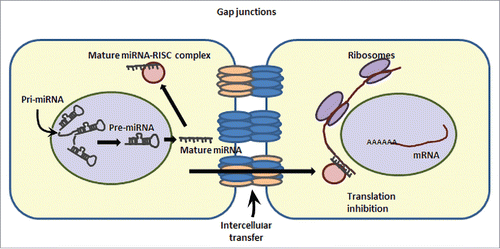
Apart from miRNA-mediated regulation of connexins, another interesting interaction exhibited by connexins/gap junctions and miRNAs involves intercellular communication via gap junction-mediated transfer of miRNAs (). With this novel mechanism, miRNAs cannot only regulate various gene transcripts within a cell of their origin but also have the ability to do so in their neighboring cells. It is yet uncertain as to what signals drive the shuttling of miRNAs between the cells via gap junctions and also whether this transfer is an active mechanism or a passive flow. Different in vitro co-culture studies have demonstrated the transfer of miRNAs between donor and recipient cells, especially in cancer cells.Citation86-89 There are no in vivo studies reported as yet, however, this specific and efficient mode of intercellular miRNA transfer would have clinical applications that include cell-specific delivery of small RNAs as a cancer therapy or for regenerative medicine. Whether such transfer exists between adjacent β-cells for regulation of insulin secretion and calcium signaling is yet unclear.
Figure 2. Intercellular transcript regulation via miRNA transfer. miRNAs can enter into neighboring cells via gap junction-mediated transfer. This cartoon demonstrates the ability of a cell (on the left) to transcribe and process the pre-miRNA to mature (single stranded) microRNA. A mature microRNA gets incorporated into RISC (RNA-induced silencing complex) and can target the expression of mRNAs in the same cell via translation inhibition/ transcript degradation or may enter the adjoining cell(s) to inhibit the expression of a specific set of genes in the neighboring cells.
Conclusion
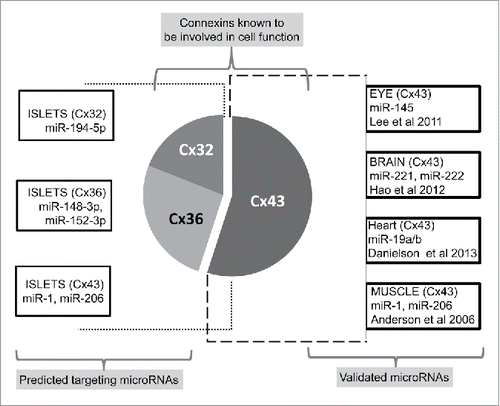
MicroRNAs have emerged as important regulators of several physiologic and pathological processes. Though most of the current literature demonstrates the role of miRNAs in Cx43 regulation (); it is most likely that such regulatory effect on other connexins will be discovered in near future. Gap junctions in pancreatic β-cells are made of Cx36; which is shown to be important for normal islet function including coordinated insulin release. The decrease in expression of Cx36 is now linked to β-cell dysfunction and prediabetes. Whether such progressive loss of Cx36 is causal to β-cell failure, leading to apoptosis, is not yet understood. Cx43 is also present in pancreatic islets but mainly in islet vasculature, which has a regulatory role in β-cell function. Given the importance of connexins in insulin secretion and the known regulation of miRs on insulin secretion (miR-375 via myotrophin and PDK1), it would be interesting to see if any of the “pancreatic” miRs regulate connexin expression and thereby insulin secretion. The potential regulation of cell-to-cell communication channels via microRNAs adds a first level of regulatory control. Interestingly, the demonstration of the regulatory molecule (dead end 1/dnd1), which can antagonize the action of these microRNAsCitation48,90 present a potentially intricate mechanism wherein such molecules could modulate connexin expression through regulation of targeting microRNAs. Further research is needed to understand if connexins, which are known to be altered in disease state, can be rescued using molecules that can either inhibit the miRNA binding or localize them to sub-cellular compartments that offer protection from miRNA-mediated degradation. If such RNA-binding proteins proffer the proposed regulatory mechanisms, then these could be used as modulators of miRNAs with potential for translational research. Studies focused on understanding the role of miRNAs in the regulation of islet-associated connexins could lead to the identification of strategies for enhancing insulin secretion in differentiating islet progenitor/stem cells.
Figure 3. Regulatory microRNAs targeting connexin gene transcripts. Schematic illustrating different microRNAs targeting the expression of specific connexin molecules demonstrated herein. The most widely studied Cx43 is experimentally shown to be targeted by several different miRNAs in various tissues (Right side panel). Left side panel represents different connexins in islets and bioinformatically identified, highly conserved (using the publically available site http://www.targetscan.org) microRNAs predicted to bind to a specific connexin.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Authors acknowledge the infrastructure support provided by the National Center for Cell Science, India, the NHMRC Clinical Trials Center, Sydney and the Rebecca L Cooper Foundation, Australia to AAH.
Funding
MRU is supported by the Department of Biotechnology, Government of India, MVJ is supported through the Australian Diabetes Society (ADS) Skip Martin Award and the JDRF International Advanced post-doctoral fellowship, WW is supported through a University of Sydney fellowship and a JDRF Australia PhD top-up scholarship and AAH is supported through JDRF Australia T1D Clinical Research Network career development award.
References
- Bosco D, Haefliger JA, Meda P. Connexins: key mediators of endocrine function. Physiol Rev 2011; 91:1393–445; PMID:22013215; https://doi.org/10.1152/physrev.00027.2010
- Dbouk HA, Mroue RM, El-Sabban ME, Talhouk RS. Connexins: a myriad of functions extending beyond assembly of gap junction channels. Cell Commun Signal 2009; 7:4; PMID:19284610; https://doi.org/10.1186/1478-811X-7-4
- Farnsworth NL, Benninger RK. New insights into the role of connexins in pancreatic islet function and diabetes. FEBS Lett 2014; 588:1278–87; PMID:24583073; https://doi.org/10.1016/j.febslet.2014.02.035
- Valiunas V, Polosina YY, Miller H, Potapova IA, Valiuniene L, Doronin S, et al. Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions. J Physiol 2005; 568:459–68; https://doi.org/10.1113/jphysiol.2005.090985
- Goodenough DA, Paul DL. Beyond the gap: functions of unpaired connexon channels. Nat Rev Mol Cell Biol 2003; 4:285–94; https://doi.org/10.1038/nrm1072
- Scemes E, Spray DC, Meda P. Connexins, pannexins, innexins: novel roles of “hemi-channels”. Pflugers Arch 2009; 457:1207–26; https://doi.org/10.1007/s00424-008-0591-5
- Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res 2004; 62:228–32; https://doi.org/10.1016/j.cardiores.2003.11.013
- Yen MR, Saier MH, Jr. Gap junctional proteins of animals: the innexin/pannexin superfamily. Prog Biophys Mol Biol 2007; 94:5–14; https://doi.org/10.1016/j.pbiomolbio.2007.03.006
- Nielsen MS, Axelsen LN, Sorgen PL, Verma V, Delmar M, Holstein-Rathlou NH. Gap junctions. Compr Physiol 2012; 2:1981–2035.
- Paul DL, Ebihara L, Takemoto LJ, Swenson KI, Goodenough DA. Connexin46, a novel lens gap junction protein, induces voltage-gated currents in nonjunctional plasma membrane of Xenopus oocytes. J Cell Biol 1991; 115:1077–89; PMID:1659572; https://doi.org/10.1083/jcb.115.4.1077
- Naus CC, Laird DW. Implications and challenges of connexin connections to cancer. Nat Rev Cancer 2010; 10:435–41; PMID:20495577; https://doi.org/10.1038/nrc2841
- Bosse Y, Despres JP, Chagnon YC, Rice T, Rao DC, Bouchard C, et al. Quantitative trait locus on 15q for a metabolic syndrome variable derived from factor analysis. Obesity (Silver Spring) 2007; 15:544–50; PMID:17372302; https://doi.org/10.1038/oby.2007.577
- Belluardo N, Trovato-Salinaro A, Mudo G, Hurd YL, Condorelli DF. Structure, chromosomal localization, and brain expression of human Cx36 gene. J Neurosci Res 1999; 57:740–52; PMID:10462698; https://doi.org/10.1002/(SICI)1097-4547(19990901)57:5%3c740::AID-JNR16%3e3.0.CO;2-Z
- Kilimnik G, Jo J, Periwal V, Zielinski MC, Hara M. Quantification of islet size and architecture. Islets 2012; 4:167–72; https://doi.org/10.4161/isl.19256
- Kim A, Miller K, Jo J, Kilimnik G, Wojcik P, Hara M. Islet architecture: A comparative study. Islets 2009; 1:129–36; https://doi.org/10.4161/isl.1.2.9480
- Steiner DJ, Kim A, Miller K, Hara M. Pancreatic islet plasticity: interspecies comparison of islet architecture and composition. Islets 2010; 2:135–45; https://doi.org/10.4161/isl.2.3.11815
- Head WS, Orseth ML, Nunemaker CS, Satin LS, Piston DW, Benninger RK. Connexin-36 gap junctions regulate in vivo first- and second-phase insulin secretion dynamics and glucose tolerance in the conscious mouse. Diabetes 2012; 61:1700–7; https://doi.org/10.2337/db11-1312
- Meda P, Chanson M, Pepper M, Giordano E, Bosco D, Traub O, et al. In vivo modulation of connexin 43 gene expression and junctional coupling of pancreatic B-cells. Exp Cell Res 1991; 192:469–80; https://doi.org/10.1016/0014-4827(91)90066-4
- Perez-Armendariz EM, Cruz-Miguel L, Coronel-Cruz C, Esparza-Aguilar M, Pinzon-Estrada E, Rancano-Camacho E, et al. Connexin 36 is expressed in beta and connexins 26 and 32 in acinar cells at the end of the secondary transition of mouse pancreatic development and increase during fetal and perinatal life. Anat Rec (Hoboken) 2012; 295:980–90; https://doi.org/10.1002/ar.22473
- Serre-Beinier V, Bosco D, Zulianello L, Charollais A, Caille D, Charpantier E, et al. Cx36 makes channels coupling human pancreatic beta-cells, and correlates with insulin expression. Hum Mol Genet 2009; 18:428–39; https://doi.org/10.1093/hmg/ddn370
- Serre-Beinier V, Le Gurun S, Belluardo N, Trovato-Salinaro A, Charollais A, Haefliger JA, et al. Cx36 preferentially connects beta-cells within pancreatic islets. Diabetes 2000; 49:727–34; PMID:10905480; https://doi.org/10.2337/diabetes.49.5.727
- Theis M, Mas C, Doring B, Kruger O, Herrera P, Meda P, et al. General and conditional replacement of connexin43-coding DNA by a lacZ reporter gene for cell-autonomous analysis of expression. Cell Commun Adhes 2001; 8:383–6; PMID:12064623; https://doi.org/10.3109/15419060109080758
- Theis M, Mas C, Doring B, Degen J, Brink C, Caille D, et al. Replacement by a lacZ reporter gene assigns mouse connexin36, 45 and 43 to distinct cell types in pancreatic islets. Exp Cell Res 2004; 294:18–29; PMID:14980497; https://doi.org/10.1016/j.yexcr.2003.09.031
- Coronel-Cruz C, Hernandez-Tellez B, Lopez-Vancell R, Lopez-Vidal Y, Berumen J, Castell A, et al. Connexin 30.2 is expressed in mouse pancreatic beta cells. Biochem Biophys Res Commun 2013; 438:772–7; https://doi.org/10.1016/j.bbrc.2013.06.100
- Peiris H, Bonder CS, Coates PT, Keating DJ, Jessup CF. The beta-cell/EC axis: how do islet cells talk to each other? Diabetes 2014; 63:3–11; PMID:24357688; https://doi.org/10.2337/db13-0617
- Benninger RK, Zhang M, Head WS, Satin LS, Piston DW. Gap junction coupling and calcium waves in the pancreatic islet. Biophys J 2008; 95:5048–61; https://doi.org/10.1529/biophysj.108.140863
- Ravier MA, Guldenagel M, Charollais A, Gjinovci A, Caille D, Sohl G, et al. Loss of connexin36 channels alters beta-cell coupling, islet synchronization of glucose-induced Ca2+ and insulin oscillations, and basal insulin release. Diabetes 2005; 54:1798–807; PMID:15919802; https://doi.org/10.2337/diabetes.54.6.1798
- Speier S, Gjinovci A, Charollais A, Meda P, Rupnik M. Cx36-mediated coupling reduces beta-cell heterogeneity, confines the stimulating glucose concentration range, and affects insulin release kinetics. Diabetes 2007; 56:1078–86; https://doi.org/10.2337/db06-0232
- Calabrese A, Zhang M, Serre-Beinier V, Caton D, Mas C, Satin LS, et al. Connexin 36 controls synchronization of Ca2+ oscillations and insulin secretion in MIN6 cells. Diabetes 2003; 52:417–24; https://doi.org/10.2337/diabetes.52.2.417
- Benninger RK, Head WS, Zhang M, Satin LS, Piston DW. Gap junctions and other mechanisms of cell-cell communication regulate basal insulin secretion in the pancreatic islet. J Physiol 2011; 589:5453–66; PMID:21930600; https://doi.org/10.1113/jphysiol.2011.218909
- Short KW, Head WS, Piston DW. Connexin 36 mediates blood cell flow in mouse pancreatic islets. Am J Physiol Endocrinol Metab 2014; 306:E324–31; https://doi.org/10.1152/ajpendo.00523.2013
- Carvalho CP, Oliveira RB, Britan A, Santos-Silva JC, Boschero AC, Meda P, et al. Impaired beta-cell-beta-cell coupling mediated by Cx36 gap junctions in prediabetic mice. Am J Physiol Endocrinol Metab 2012; 303:E144–51; https://doi.org/10.1152/ajpendo.00489.2011
- Porte D, Jr., Kahn SE. beta-cell dysfunction and failure in type 2 diabetes: potential mechanisms. Diabetes 2001; 50 Suppl 1:S160–3; PMID:11272181; https://doi.org/10.2337/diabetes.50.2007.S160
- Urschel S, Hoher T, Schubert T, Alev C, Sohl G, Worsdorfer P, et al. Protein kinase A-mediated phosphorylation of connexin36 in mouse retina results in decreased gap junctional communication between AII amacrine cells. J Biol Chem 2006; 281:33163–71; PMID:16956882; https://doi.org/10.1074/jbc.M606396200
- Farnsworth NL, Walter RL, Hemmati A, Westacott MJ, Benninger RK. Low Level Pro-inflammatory Cytokines Decrease Connexin36 Gap Junction Coupling in Mouse and Human Islets through Nitric Oxide-mediated Protein Kinase Cdelta. J Biol Chem 2016; 291:3184–96; PMID:26668311; https://doi.org/10.1074/jbc.M115.679506
- Penko D, Rojas-Canales D, Mohanasundaram D, Peiris HS, Sun WY, Drogemuller CJ, et al. Endothelial progenitor cells enhance islet engraftment, influence beta-cell function, and modulate islet connexin 36 expression. Cell Transplant 2015; 24:37–48; PMID:24069942; https://doi.org/10.3727/096368913X673423
- Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res 2006; 34:5863–71; https://doi.org/10.1093/nar/gkl743
- Hao J, Zhang C, Zhang A, Wang K, Jia Z, Wang G, et al. miR-221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol Rep 2012; 27:1504–10.
- Hsieh YW, Chang C, Chuang CF. The microRNA mir-71 inhibits calcium signaling by targeting the TIR-1/Sarm1 adaptor protein to control stochastic L/R neuronal asymmetry in C. elegans. PLoS Genet 2012; 8:e1002864; https://doi.org/10.1371/journal.pgen.1002864
- Jin Z, Xu S, Yu H, Yang B, Zhao H, Zhao G. miR-125b inhibits Connexin43 and promotes glioma growth. Cell Mol Neurobiol 2013; 33:1143–8; https://doi.org/10.1007/s10571-013-9980-1
- Joglekar MV, Parekh VS, Hardikar AA. New pancreas from old: microregulators of pancreas regeneration. Trends Endocrinol Metab 2007; 18:393–400; PMID:18023200; https://doi.org/10.1016/j.tem.2007.10.001
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97; PMID:14744438; https://doi.org/10.1016/S0092-8674(04)00045-5
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642–55; PMID:19239886; https://doi.org/10.1016/j.cell.2009.01.035
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–33; https://doi.org/10.1016/j.cell.2009.01.002
- Tufekci KU, Oner MG, Meuwissen RL, Genc S. The role of microRNAs in human diseases. Methods Mol Biol 2014; 1107:33–50; PMID:24272430.
- Tufekci KU, Meuwissen RL, Genc S. The role of microRNAs in biological processes. Methods Mol Biol 2014; 1107:15–31; PMID:24272429.
- Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 2009; 7:147–54; PMID:20172487; https://doi.org/10.1016/S1672-0229(08)60044-3
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007; 131:1273–86; PMID:18155131; https://doi.org/10.1016/j.cell.2007.11.034
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, et al. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol 2003; 13:1429–34; PMID:12932328; https://doi.org/10.1016/S0960-9822(03)00537-2
- Dumortier O, Van Obberghen E. MicroRNAs in pancreas development. Diabetes Obes Metab 2012; 14 Suppl 3:22–8; PMID:22928561; https://doi.org/10.1111/j.1463-1326.2012.01656.x
- Guay C, Jacovetti C, Nesca V, Motterle A, Tugay K, Regazzi R. Emerging roles of non-coding RNAs in pancreatic beta-cell function and dysfunction. Diabetes Obes Metab 2012; 14 Suppl 3:12–21; PMID:22928560; https://doi.org/10.1111/j.1463-1326.2012.01654.x
- Joglekar MV, Parekh VS, Hardikar AA. Islet-specific microRNAs in pancreas development, regeneration and diabetes. Indian J Exp Biol 2011; 49:401–8; PMID:21702218.
- Joglekar MV, Joglekar VM, Hardikar AA. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr Patterns 2009; 9:109–13; PMID:18977315; https://doi.org/10.1016/j.gep.2008.10.001
- Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A 2009; 106:5813–8; PMID:19289822; https://doi.org/10.1073/pnas.0810550106
- Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic beta-cells. Diabetes 2013; 62:887–95; PMID:23223022; https://doi.org/10.2337/db12-0451
- Nieto M, Hevia P, Garcia E, Klein D, Alvarez-Cubela S, Bravo-Egana V, et al. Antisense miR-7 impairs insulin expression in developing pancreas and in cultured pancreatic buds. Cell Transplant 2012; 21:1761–74; https://doi.org/10.3727/096368911X612521
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004; 432:226–30; PMID:15538371; https://doi.org/10.1038/nature03076
- El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3'-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 2008; 57:2708–17; PMID:18591395; https://doi.org/10.2337/db07-1614
- Tattikota SG, Sury MD, Rathjen T, Wessels HH, Pandey AK, You X, et al. Argonaute2 regulates the pancreatic beta-cell secretome. Mol Cell Proteomics 2013; 12:1214–25; https://doi.org/10.1074/mcp.M112.024786
- Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 2010; 59:978–86; PMID:20086228; https://doi.org/10.2337/db09-0881
- Ruan Q, Wang T, Kameswaran V, Wei Q, Johnson DS, Matschinsky F, et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc Natl Acad Sci U S A 2011; 108:12030–5; PMID:21730150; https://doi.org/10.1073/pnas.1101450108
- Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes 2008; 57:2728–36; PMID:18633110; https://doi.org/10.2337/db07-1252
- Roggli E, Gattesco S, Caille D, Briet C, Boitard C, Meda P, et al. Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes 2012; 61:1742–51; PMID:22537941; https://doi.org/10.2337/db11-1086
- Farr RJ, Januszewski AS, Joglekar MV, Liang H, McAulley AK, Hewitt AW, et al. A comparative analysis of high-throughput platforms for validation of a circulating microRNA signature in diabetic retinopathy. Sci Rep 2015; 5:10375; https://doi.org/10.1038/srep10375
- Joglekar MV, Januszewski AS, Jenkins AJ, Hardikar AA. Circulating microRNA Biomarkers of Diabetic Retinopathy. Diabetes 2016; 65:22–4; https://doi.org/10.2337/dbi15-0028
- Zhu H, Leung SW. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia 2015; 58:900–11; https://doi.org/10.1007/s00125-015-3510-2
- Zhao X, Mohan R, Ozcan S, Tang X. MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic beta-cells. J Biol Chem 2012; 287:31155–64; https://doi.org/10.1074/jbc.M112.362632
- Sebastiani G, Po A, Miele E, Ventriglia G, Ceccarelli E, Bugliani M, et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol 2015; 52:523–30; https://doi.org/10.1007/s00592-014-0675-y
- Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem 2008; 389:305–12; https://doi.org/10.1515/BC.2008.026
- Wijesekara N, Zhang LH, Kang MH, Abraham T, Bhattacharjee A, Warnock GL, et al. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes 2012; 61:653–8; https://doi.org/10.2337/db11-0944
- Laird DW. Life cycle of connexins in health and disease. Biochem J 2006; 394:527–43; PMID:16492141; https://doi.org/10.1042/BJ20051922
- Klotz LO. Posttranscriptional regulation of connexin-43 expression. Arch Biochem Biophys; 524:23–9; PMID:22464988; https://doi.org/10.1016/j.abb.2012.03.012
- Zhou R, Hang P, Zhu W, Su Z, Liang H, Du Z. Whole genome network analysis of ion channels and connexins in myocardial infarction. Cell Physiol Biochem 2011; 27:299–304; https://doi.org/10.1159/000327956
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med 2007; 13:486–91; PMID:17401374; https://doi.org/10.1038/nm1569
- Lu Y, Zhang Y, Shan H, Pan Z, Li X, Li B, et al. MicroRNA-1 downregulation by propranolol in a rat model of myocardial infarction: a new mechanism for ischaemic cardioprotection. Cardiovasc Res 2009; 84:434–41; PMID:19581315; https://doi.org/10.1093/cvr/cvp232
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 2006; 174:677–87; https://doi.org/10.1083/jcb.200603008
- Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci U S A 2009; 106:20794–9; https://doi.org/10.1073/pnas.0909311106
- Curcio A, Torella D, Iaconetti C, Pasceri E, Sabatino J, Sorrentino S, et al. MicroRNA-1 downregulation increases connexin 43 displacement and induces ventricular tachyarrhythmias in rodent hypertrophic hearts. PLoS One 2013; 8:e70158; https://doi.org/10.1371/journal.pone.0070158
- Imamura M, Sugino Y, Long X, Slivano OJ, Nishikawa N, Yoshimura N, et al. Myocardin and microRNA-1 modulate bladder activity through connexin 43 expression during post-natal development. J Cell Physiol 2013; 228:1819–26; https://doi.org/10.1002/jcp.24333
- Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res 2011; 71:2381–91; https://doi.org/10.1158/0008-5472.CAN-10-2754
- Lee SK, Teng Y, Wong HK, Ng TK, Huang L, Lei P, et al. MicroRNA-145 regulates human corneal epithelial differentiation. PLoS One 2011; 6:e21249; https://doi.org/10.1371/journal.pone.0021249
- Li X, Pan JH, Song B, Xiong EQ, Chen ZW, Zhou ZS, et al. Suppression of CX43 expression by miR-20a in the progression of human prostate cancer. Cancer Biol Ther 2012; 13:890–8; https://doi.org/10.4161/cbt.20841
- Danielson LS, Park DS, Rotllan N, Chamorro-Jorganes A, Guijarro MV, Fernandez-Hernando C, et al. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J 2013; 27:1460–7; https://doi.org/10.1096/fj.12-221994
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009; 119:2772–86; PMID:19726871; https://doi.org/10.1172/JCI36154
- Wang BW, Wu GJ, Cheng WP, Shyu KG. Mechanical stretch via transforming growth factor-beta1 activates microRNA-208a to regulate hypertrophy in cultured rat cardiac myocytes. J Formos Med Assoc 2013; 112:635–43; PMID:24120154; https://doi.org/10.1016/j.jfma.2013.01.002
- Hong X, Sin WC, Harris AL, Naus CC. Gap junctions modulate glioma invasion by direct transfer of microRNA. Oncotarget 2015; 6:15566–77; https://doi.org/10.18632/oncotarget.3904
- Lemcke H, Steinhoff G, David R. Gap junctional shuttling of miRNA - A novel pathway of intercellular gene regulation and its prospects in clinical application. Cell Signal 2015; 27:2506–14; https://doi.org/10.1016/j.cellsig.2015.09.012
- Lim PK, Bliss SA, Patel SA, Taborga M, Dave MA, Gregory LA, et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res 2011; 71:1550–60; https://doi.org/10.1158/0008-5472.CAN-10-2372
- Suzhi Z, Liang T, Yuexia P, Lucy L, Xiaoting H, Yuan Z, et al. Gap Junctions Enhance the Antiproliferative Effect of MicroRNA-124-3p in Glioblastoma Cells. J Cell Physiol 2015; 230:2476–88; https://doi.org/10.1002/jcp.24982
- Ketting RF. A dead end for microRNAs. Cell 2007; 131:1226–7; PMID:18160032; https://doi.org/10.1016/j.cell.2007.12.004

