ABSTRACT
The conserved glycosylation site Asn297 of a monoclonal antibody (mAb) can be decorated with a variety of sugars that can alter mAb pharmacokinetics and recruitment of effector proteins. Antibodies lacking the core fucose at Asn297 (afucosylated mAbs) show enhanced antibody-dependent cell-mediated cytotoxicity (ADCC) and increased efficacy. Here, we describe the development of a robust platform for the manufacture of afucosylated therapeutic mAbs by engineering a Chinese hamster ovary (CHO) host cell line to co-express a mAb with GDP-6-deoxy-D-lyxo-4-hexulose reductase (RMD), a prokaryotic enzyme that deflects an intermediate in the de novo synthesis of fucose to a dead-end product, resulting in the production of afucosylated mAb (GlymaxX™ Technology, ProBioGen). Expression of the mAb and RMD genes was coordinated by co-transfection of separate mAb and RMD vectors or use of an internal ribosome entry site (IRES) element to link the translation of RMD with either the glutamine synthase selection marker or the mAb light chain. The GS-IRES-RMD vector format was more suitable for the rapid generation of high yielding cell lines, secreting afucosylated mAb with titers exceeding 6.0 g/L. These cell lines maintained production of afucosylated mAb over 60 generations, ensuring their suitability for use in large-scale manufacturing. The afucosylated mAbs purified from these RMD-engineered cell lines showed increased binding in a CD16 cellular assay, demonstrating enhancement of ADCC compared to fucosylated control mAb. Furthermore, the afucosylation in these mAbs could be controlled by simple addition of L-fucose in the culture medium, thereby allowing the use of a single cell line for production of the same mAb in fucosylated and afucosylated formats for multiple therapeutic indications.
Introduction
Recombinant Chinese hamster ovary (CHO) cells are the industry standard platform for large-scale production and commercialization of human therapeutic proteins.Citation1,Citation2 The robust growth, high productivity and stability of CHO cell lines enable consistency in the bioprocesses essential for the manufacture of biologic drugs. Glycoproteins produced in CHO cells contain a variety of simple and complex glycan structures,Citation3 and some glycans have significant effects on the properties of therapeutically administered proteins.Citation4-7 Of particular importance is the presence or absence of terminal sialic acid,Citation8-10 which affects the clearance and therefore circulatory half-life of biotherapeutic proteins. Antibodies normally do not contain significant levels of sialic acid unless glycosylation occurs at sites other than Asn297, which is found in the constant region of IgG.Citation6,Citation11 This site is protected and does not have significant levels of terminal sialic acid.Citation12 However, other modifications of this site, in particular the presence of high mannose glycoforms and the presence or absence of fucose, can modulate antibody effector function. The glycoforms present at Asn297 influence biological activity by mediating antibody-dependent cell-mediated cytotoxicity (ADCC),Citation7,Citation13-15 a critically important function for therapeutic mAbs directed against infections and tumor cells.Citation16,Citation17 An antibody bound to membrane-surface antigens of a target cell triggers classical ADCC, mediated by recruitment of the natural killer (NK) cells, macrophages, neutrophils and eosinophils.Citation17 The ADCC response is activated when the CD16 receptors (FcγRIII) on the NK cells bind to the Fc region of an IgG molecule,Citation18 which induces the directional release of cytolytic mediators, such as perforin, as well as enzymes including granzyme-B, which enter a targeted cell to set off a cascade of events that result in apoptosis and killing of the target cell.Citation19
Increased ADCC activity may allow reduced dose requirements for a therapeutic mAb,Citation20 and so provides the potential for reduced cost of goods if the mAb can be manufactured efficiently.Citation21 Various strategies using CHO cell line engineering or process development have been employed to modulate the glycoforms that affect ADCC. It has been shown that co-expression of glycosylation enzyme β1-4-N-acytylglucosaminyltransferease III (GNTIII) in CHO cells introduced bisected GlcNAC to secreted anti-CD20 mAb, which exhibited increased potency by enhanced binding to Fc gamma RIII-expressing cells.Citation22 On the other hand, antibodies containing high levels of N-linked mannose-5 glycan (Man5) have been reported to exhibit enhanced ADCC compared with antibodies with fucosylated complex or hybrid glycans.Citation23 Highly galactosylated glycostructures on therapeutic mAbs exhibit increased in vitro C1q-binding and complement-dependent cytotoxicity (CDC) activity.Citation24,Citation25 Increased galactosylation of CHO-derived antibodies has been shown to increase FcγRII and FcγRIIIa bindingCitation26 and enhanced ADCC activity.Citation27 However, the effect of galactosylation is subtle compared with the effect of afucosylation.Citation26
Although mAbs lacking the core fucose show enhanced ADCC activity,Citation15,Citation28-35 most mAbs generated in CHO cells contain α-1-6 fucosylation. Since CHO-expressed mAbs contain 90% fucosylated carbohydrates, several strategies have been devised to reduce or eliminate fucosylation in CHO cells.Citation36,Citation37 It has been successfully shown that afucosylated mAbs can be generated from CHO cells by knocking down, or knocking out the FUT8 gene, which encodes α1,6-fucosyltransferase that catalyzes the transfer of fucose from GDP-fucose to N-acetylglucosamine (GlcNAc) in an α-1,6 linkage.Citation38,Citation39 Completely non-fucosylated antibodies can also be generated in CHO-DG44 hosts by knocking down expression of GDP-fucose 4,6-dehydratase (GMD).Citation40 Additionally, the fucosylation of mAbs produced by CHO cells can be altered by supplementing the culture medium with an inhibitor of glycosyl transferase, such as 2F-peracetyl-fucose.Citation41
Recently, it was shown that heterologous expression of a bacterial oxidoreductase GDP-6-deoxy-D-lyxo-4-hexulose reductase (RMD) provides a reliable method for conversion of an existing antibody-producing cell line into a cell line capable of secreting afucosylated IgG lacking the core fucose moiety.Citation42 Since fucose is not typically present in CHO expression medium, cells depend on de novo generation of fucose. RMD functions as a deflecting enzyme to block the fucosylation pathway by enzymatic conversion of GDP-4-keto-6-deoxymannose, a metabolic intermediate of the pathway, to GDP-D-Rhamnose, a dead-end metabolite and a sugar that cannot be metabolized by CHO cells.Citation42,Citation43 In previously published work, an existing mAb-producing cell line was engineered to express RMD or an RMD-expressing CHO cell line was engineered to express a mAb.Citation43 Both approaches involved two rounds of transfection, selection and screening. Here, we report the development of a simplified, single-step method for the rapid generation of CHO cell lines producing afucosylated mAbs using RMD co-expression. This strategy uses an existing CHO host cell line, delivering cell lines that are compatible with established upstream platform processes, scalable for manufacturing and suitable for commercialization.
Results
Generation of stable IgG cell lines co-expressing RMD
Expression of RMD in IgG-producing cells has already been shown to be an effective way of producing afucosylated IgG.Citation42,Citation43 In an effort to streamline the cell line generation for the production of afucosylated IgGs, we constructed a set of plasmids for the co-expression of IgG and RMD. To evaluate the best expression strategy, three plasmids were generated (). In one, RMD was placed directly under control of a CMV promoter in a vector independent of the IgG expression vector (CMV-RMD). In the other two vectors, the RMD cassette was cloned after an IRES sequence following either the glutamine synthase (GS) gene (, GS-IRES-RMD) with transcription driven by the SV40 promoter, or following the IgG light chain (LC) gene (LC-IRES-RMD) with transcription driven by the CMV promoter. A scheme of the cell line isolation and characterization is summarized in . Following transfection and selection, colonies from the GS-IRES-RMD and control IgG vectors (-RMD) showed similar hit rates for positive IgG expressers (51% and 45%, respectively; ). A much lower number (17%) of the LC-IRES-RMD colonies expressed IgG. Interestingly, all the colonies derived from the co-transfection of RMD and IgG plasmids showed IgG expression, and this may reflect the increased stringency of the dual selection agents.
Figure 1. Cell line development for co-expression of RMD and IgG for generation of afucosylated mAb. A. Schematic representation of expression plasmids for RMD and IgG HC and IgG LC, either in two separate vectors or in single vectors (GS-IRES-RMD and LC-IRESRMD). B. Strategy for cell line engineering and screening to co-express IgG and RMD for production of afucosylated mAb. Ci. Distribution of geometric means for stable cell lines surface-stained with FITC-LCA. LCA-stained cells from -RMD, +RMD, GS-IRES-RMD and LC-IRES-RMD were analyzed using LSRII instrument. Cii. Distribution of geometric means for stable cell lines surface-stained with FITC-LCA. LCA-stained cells from co-transfection (RMD and IgG) were analyzed using FACSCalibur instrument. Unpaired t test was used to determine the P values. D. IgG Titer distribution of stable cell lines expressing fucosylated (-RMD) or afucosylated mAbs derived from different expression vectors. Unpaired t test was used to determine the P values.
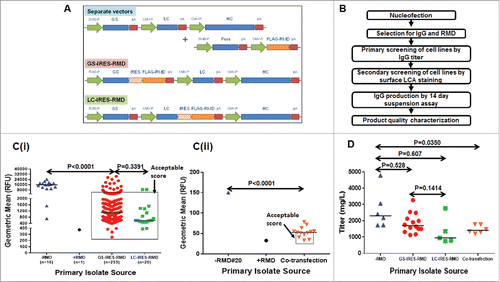
Table 1. Colony screening summary.
The effect of RMD expression on IgG fucosylation was evaluated using a fluorescently labeled lentil lectin, Lens culinaris agglutinin (LCA). LCA specifically binds to N-linked glycan structures containing α1-6 fucose. Thus, cells expressing afucosylated IgG should show reduced LCA binding. Indeed, cell lines expressing RMD showed a significant decrease in staining (C). While the difference in the median geometric mean of LCA-stained cells from the GS-IRES-RMD and LC-IRES-RMD constructs were not statistically significant (p = 0.3391), cells from both constructs showed a significant decrease in LCA staining relative to IgG stable cell lines not expressing RMD (-RMD) (p < 0.001). However, with the GS-IRES-RMD construct, a large number of isolates retained high LCA binding indicating that RMD levels were not optimal in all isolates.
To further examine cells with low LCA staining, we established an arbitrary cut off for geometric means of 5000 relative fluorescence units (RFU), measured in an LSRII instrument (BD Biosciences). Cells within this gate were considered to have an ‘acceptable score’ and were selected for further evaluation in a fed-batch assay for IgG expression (Ci). For the cell lines generated by co-transfection, the LCA-stained cells were analyzed using a FACSCalibur (BD Biosciences) instrument. A lower threshold of geometric mean (<50) was set to select these isolates for IgG expression evaluation (Cii). It is noteworthy that the cut off value for geometric mean depends on several factors such as voltage, laser strength, and the sensitivity of the flow cytometer. For instance, LSRII can analyze less than 80 molecules of equivalent soluble fluorochrome, whereas the much less sensitive FACSCalibur instrument can analyze 750 molecules of equivalent soluble fluorochromes. A stable IgG-expressing cell line co-expressing RMD was used as a positive control for LCA staining and represented as +RMD.Citation42
The cell lines with low LCA binding were evaluated for IgG expression in multi-well format (24-deep-well plates). In addition, the top expressers from each group were re-evaluated for IgG expression in shake flasks (D). A slightly higher median titer was seen among the cell lines transfected with the IgG cassettes alone (2.2 g/L) compared to the GS-IRES-RMD cell lines (1.8 g/L). However, the difference was not statistically significant (p = 0.528). In contrast, the median titer for the primary isolates derived from the LC-IRES-RMD vector was much lower (<1.0 g/L) compared to the RMD negative IgG cassette alone and the GS-IRES-RMD vector. Titers for co-transfection cell lines were much less diverse, with a median titer of 1.3 g/L (D). The data suggests that RMD did not have a negative impact on IgG expression.
To characterize the observed differences in the production of afucosylated IgG, western blot analysis of whole cell extracts from cell lines containing the GS-IRES-RMD and LC-IRES-RMD constructs were performed to detect RMD protein (). We observed variable expression of RMD from the GS-IRES-RMD construct across a number of primary isolates. The lack of RMD expression in many of these isolates explains why many colonies with this construct expressed fucosylated mAb (Ci). In contrast, robust and homogeneous RMD protein expression was observed for the LC-IRES-RMD vector (). A similar profile for RMD expression was observed by qPCR (). The differential RMD expression for these two constructs was perhaps due to the difference in promoters driving transcription, i.e., the SV40 promoter for the GS-IRES-RMD construct compared with the ‘stronger’ CMV promoter for the LC-IRES-RMD construct. However, the difference in RMD expression was independent of GS expression levels because the GS protein levels remained equivalent across the isolates for both GS-IRES-RMD and LC-IRES-RMD vectors (C).
Figure 2. Biochemical characterization of stable cell lines and secreted IgG products. A. Detection of intracellular FLAG-RMD by western blotting in cell lysates extracted from stable primary isolates derived from GS-IRES-RMD, LC-IRES-RMD and -RMD control vectors. B. Detection of RMD mRNA expression by QRTPCR in cell lines derived from GS-IRES-RMD, LC-IRES-RMD and -RMD vectors. C. Detection of GS expression by western blotting in cell lysates extracted from cell lines derived from GS-IRES-RMD, LC-IRES-RMD and -RMD vectors. D. N-linked glycan profiles of the purified IgG products derived from cell lines with high RMD expression (GS-IRES-RMD#14, LC-IRES-RMD #15 and #17), intermediate RMD expression (GS-IRES-RMD (#2, #3) and low RMD expression (GS-IRES-RMD #6). E. Competitive cell based HTRF assay monitoring Fc gamma receptor binding (CD16a) of IgG purified from selected RMD cell lines.
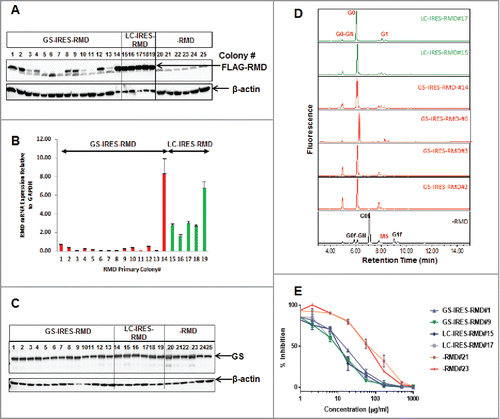
In order to examine the effect of differences in RMD expression levels on the degree of IgG afucosylation, the N-linked glycans were released from purified IgG by PNGase F digestion and analyzed by mass spectrometry (MS). IgG samples were assessed from GS-IRES-RMD cell lines expressing high (isolates #2 and #14), intermediate (isolate #3) and low (isolate #6) levels of RMD, as well as from LC-IRES-RMD cell lines expressing high levels of RMD (#15 and #17) and control cell lines not expressing RMD (-RMD). Our data show that expression of RMD at any level results in the virtual elimination of fucosylation (D and ). G0f and G0f-GN forms of the sugar moiety were identified only in IgG from cell lines not expressing RMD. It is evident from the data that the core fucose groups were significantly lower in abundance, or even below the detection level, in isolates derived from both GS-IRES-RMD and LC-IRES-RMD groups ( and ). The complete release of the glycosylation group was confirmed by intact mass analysis of PNGaseF treated samples (Supplemental Fig. 1.)
Table 2a. Fucosylation status and glycosylation profile of mAbA in presence or absence of RMD.
The effector function for the IgG from the RMD-expressing cell lines was determined using a HTRF-based CD16a cellular binding assay. Afucosylated IgG shows higher affinity for FcγRIIIa (CD16a), which correlates with improved ADCC.Citation44 CD16a is a multi-chain immune recognition receptor composed of a binding chain and signaling gamma chain.Citation45 In this assay, HEK293 cells co-expressing the CD16a receptor and the gamma signaling chain were labeled with a Terbium donor dye. Binding of human IgG labeled with d2 acceptor to the CD16a receptor results in a FRET signal. Addition of purified IgG from GS-IRES-RMD and LC-IRES-RMD isolates to these cells competes with IgG-d2 for binding to the CD16a receptor and results in loss of FRET signal, which is inversely proportional to the IgG affinity. IgG from GS- and LC-IRES-RMD isolates, which lacked the core fucose moiety, showed higher binding affinity for CD16a compared to RMD negative isolates 21 and 23 (E). Likewise, EC50 values for the IgG from RMD negative isolates were 5-8-fold higher than IgG derived from the RMD-expressing cell lines (). However, an earlier report suggested that levels of afucosylated glycans influence ADCC in a linear fashion.Citation46 Taken together, the data suggest that RMD is a highly active enzyme that does not need to be expressed at high levels in order to substantially reduce fucosylation of IgGs, which allows enhanced binding to CD16 receptors (, , and ).
Table 2b. Determination of EC50 in cell-based potency assay.
Application of the GS-IRES-RMD vector format for IgG Expression in a stable cell line development campaign
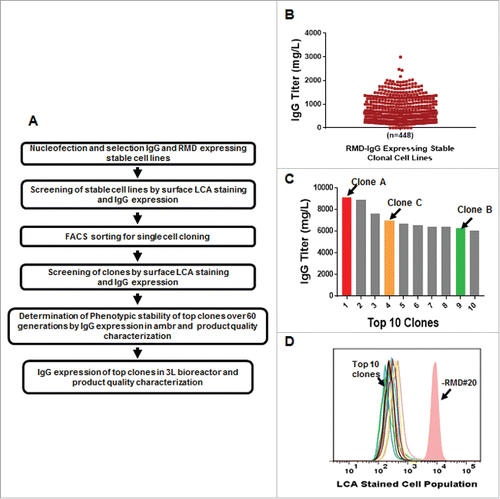
After successful evaluation of the different expression constructs, the GS-IRES-RMD vector format was chosen to engineer CHO cells, which had been derived from CHOK1 by adapting the cells to growth in suspension in medium, for expression of a candidate therapeutic mAb. Even though the RMD expression levels were markedly different between GS-IRES-RMD and LC-IRES-RMD, both formats offered a similar degree of afucosylation of the secreted IgG product. However, an equivalent colony outgrowth efficiency and IgG productivity compared to conventional IgG format positioned GS-IRES-RMD to be the preferred format for generation of candidate therapeutic mAbs. A schematic workflow for the cell screening and characterization that includes a long-term stability study is summarized in . IgG-expressing stable cell lines with low LCA binding were pooled and sorted as single cells in 384-well plates by fluorescence-activated cell sorting (FACS).Citation47 A total of 448 clones were screened in 96 deep-well fed-batch cultures for IgG expression, with the top clone reaching a titer of 2.5 g/L on day 10 (). The fucosylation status of the top 110 clones, ranked based on day 10 titers, were further evaluated by LCA staining. A more controlled IgG titer evaluation, with frequent monitoring of cell growth and metabolism over the course of 14-day fed-batch was then performed in shake flasks for the top 10 clones that were high in IgG titer and low in LCA binding, followed by a round of LCA cell surface staining for a qualitative assessment of afucosylation status. The titers for all 10 clones exceeded 6.0 g/L with the top clone reaching 9.0 g/L (C). All 10 clones showed substantially lower LCA binding than a clone not expressing RMD (-RMD#20) (D).
Figure 3. Application of the GS-IRES-RMD Format for IgG Expression in Stable Cell Line Development Campaign. A. Strategy for cell line engineering, clone selection and phenotypic stability determination for co-expressing IgG and RMD in a single vector format for production of afucosylated mAb. B. Titer distribution on day 10 of mAb expressing clonal cells screened in 96-deep-well plates in high throughput format. C. Titer distribution of top 10 clones in 14-day fed-batch assay in shake flasks. D. LCA staining showing low binding due to afucosylation of top 10 clones. -RMD#20 cell line was used as a positive control for LCA binding to cell surface.
Three clones, A, B and C, with high IgG titer and low LCA surface binding were chosen as candidates for a long-term phenotypic stability study. The three clones were routinely passaged for 60 generations and cells were cryo-preserved at 10, 30, 45 and 60 population doublings (PDLs). The cryopreserved cells were revived and then simultaneously evaluated for cell growth, metabolism and IgG production in duplicate in an automated microscale bioreactor system (ambr15™). At harvest, IgG was also purified for product quality characterization. In addition to maintaining consistent IgG expression throughout 60 generations (), all three clones showed low fucosylation status as determined by LCA surface staining (). The three clones were further evaluated in bench-scale 3 L bioreactors at 45 PDLs to compare with the performance in ambr15™. The results indicate consistent IgG expression levels for the selected individual clones from milliliter scale () to bench-top bioreactor scale (C). The clones also maintained consistent afucosylation status during the phenotypic stability studies, when the cell culture supernatants were analyzed between 10 and 60 PDLs in milliliter scale ambr15™ bioreactors, and compared with purified material generated in 3 L bioreactors at 45 PDL (). The results show consistent glycan profiles across the early- and late-passage cells and between small and scaled up bioreactor formats, and the fucosylated species remained below the limit of -quantification (<LOQ = 1%) as well ().
Figure 4. Phenotypic stability studies for RMD-IgG Clones in GS-IRES-RMD format. A. Consistent IgG production profile of RMD-IgG Clones A, B and C at 10, 30, 45 and 60 PDLs performed in milliliter scale in ambr15™ bioreactors in duplicate. B. Evaluation of LCA binding, a metric of afucosylation, to cell surface of IgG expressing clones co-expressing RMD at 10, 30, 45 and 60 PDL. C. Consistency in IgG expression in RMD-IgG Clones A, B and C at 45 PDL in milliliter scale in ambr15™ bioreactors and platform bench-scale 3L bioreactors.
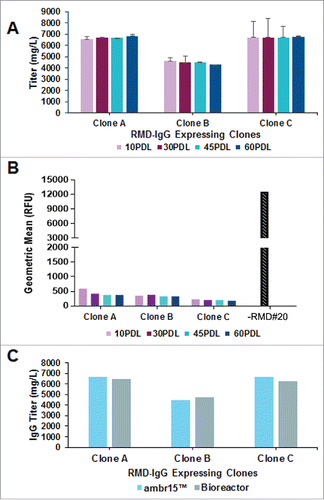
Table 3. Fucosylation status and glycosylation profile for 3 clones co-expressing RMD.
Controlled fucosylation of mabs in RMD cell lines
von Hornsten et al.Citation42 have speculated that the deflecting activity of RMD may be bypassed by the addition of L-fucose into the culture medium, thereby replenishing the cytosolic GDP-L-fucose and restoring the fucosylation of secreted IgG. The flexibility of switching an IgG molecule between its fucosylated and afucosylated forms by supplementing fucose has an enormous advantage for manufacturing the same IgG molecule for different therapeutic indications requiring diverse effector functions. To test whether the fucosylation state is switchable in this system, we generated IgG samples from representative GS-IRES-RMD, LC-IRES-RMD and -RMD isolates by culturing cells in growth medium supplemented with different concentrations of L-fucose (0, 5, 10, 20, 50 and 100 µM). IgG purified from these cells was tested for fucosylation levels using biochemical as well as cell-based assays. A gradual shift in the cell population with increasing concentrations of L-fucose was noticeable by flow cytometry due to the conversion from LCA negative to LCA positive staining influenced by the change in fucosylation status assessed on day 6 of the fed-batch assay (). The LCA binding peaked in the presence of 50 µM L-fucose and did not increase further in the presence of 100 µM L-fucose concentration in both GS-IRES-RMD#6 and LC-IRES-RMD#17 primary isolates ( and ). However, cell surface binding of LCA remained constant in the absence (0 µM) or presence (100 µM) of L-fucose in the RMD-negative cell line -RMD#20. As the cells became incrementally positive for LCA surface binding when GS-IRES-RMD and LC-IRES-RMD cell lines were cultured in presence of increasing concentrations of L-fucose, the total fucosylation also increased in a linear fashion with R-squared value of 1.00 for GS-IRES-RMD#6 and 0.94 for LC-IRES_RMD#17 with increasing concentrations of L-fucose and a nearly complete conversion to fucosylated form of IgG was observed in cells cultured in the presence of 100 µM L-fucose (C, Supplemental Fig. 2 and and ). The fucosylation levels remained unchanged for RMD-negative cell line -RMD#20 in the presence or absence of L-fucose in culture medium (, (C). Although, maximal LCA staining is observed at 50 µM L-fucose, fucosylation is still higher at 100 µM ( and Supplemental Fig. 2). Interestingly, cells with an LCA geometric mean of lower than 5000 RFU are capable of producing fully afucosylated mAbs, which supports the previously described threshold (Ci).
Figure 5. Reversal of afucosylation of secreted mAbs by addition of L-fucose to cell culture medium. Stable IgG cell lines co-expressing RMD were cultured in the presence or absence of L-fucose and the IgG produced monitored for fucosylation. A. Flow cytometry analysis of LCA stained cells from GS-IRES-RMD#6, LC-IRES-RMD#17 and -RMD#20 cell lines on day 6 of fed-batch culture in the presence of different concentrations of L-fucose (0, 5, 10, 20, 50, 100 µM). B. Statistical analyses of geometric mean derived from LCA cell surface stained cells growing in presence of 0, 5, 10, 20, 50 or, 100 µM of L-fucose. C. Glycan profiles of IgG purified from primary isolates GS-IRES-RMD#6 and LC-IRES-RMD#17 grown in presence of different concentrations of L-fucose. D. Western Blot analysis of purified IgG from cells expressing RMD using biotinylated LCA to detect the fucose moiety (f-HC) (upper panel). The membrane was probed for IgG HC and LC to confirm equal sample loading (lower panel). E and F. Activity of the IgG samples purified from primary isolate GS-IRES-RMD#6, LC-IRES-RMD#17 and -RMD#20 cultured in presence of different concentrations of L-fucose was measured by an HTRF-based CD16a cellular binding assay. afucosylated antibodies from GS-IRES-RMD#6 and LC-IRES-RMD#17 cell lines cultured in 0–50 µM L-fucose (dotted lines) showed higher binding activity to CD16a than fucosylated antibodies purified from cells cultured in presence of 100 µM L-fucose (solid lines).
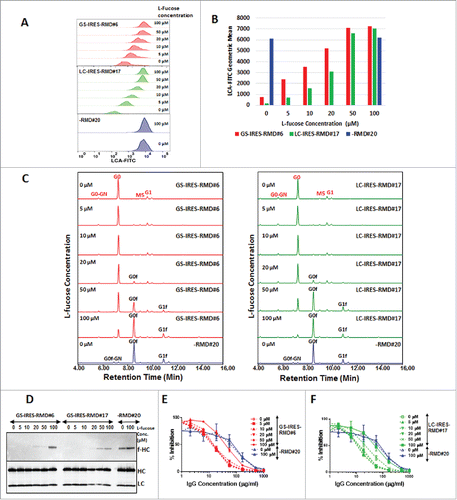
Table 4a. (i). Reversal of afucosylation by addition of fucose in cell culture medium for RMD expressing primary isolate GS-IRES-RMD#6.
Table 4a. (ii). Reversal of afucosylation by addition of fucose in cell culture medium for RMD expressing primary isolate LC-IRES-RMD#17.
Table 4a. (iii). Reversal of afucosylation by addition of fucose in cell culture medium for RMD negative primary isolate -RMD#20.
The change in mass of the mAb due to fucosylation was further confirmed by intact mass analysis (Supplemental Fig. 3). Fucosylation of the heavy chain (HC) by the influence of L-fucose in the culture medium was also detected by western blot analysis of purified mAbs using the biotinylated LCA, where fucosylated HC was detected in IgG samples derived from cells treated with as little as 20 µM L-fucose (D). Additionally, the HTRF-based CD16 assay in HEK293 cells confirmed the loss of binding activity of the fucosylated IgG compared to the original afucosylated IgG in GS-IRES-RMD#6 and LC-IRES-RMD#17 (E and F). Likewise, a concentration-dependent increase in EC-50 was observed more prominently in LC-IRES-RMD#17 in the presence of 50 µM L-fucose, at which concentration partial fucosylation of IgG was detected (). The EC-50 values increased further with the switch to fucosylated IgG that occurred in the presence of 100 µM L-fucose in the culture medium (). These data suggest that when fucose is added in the culture medium, fucosylation occurs by modulating the cytosolic GDP L-fucose pools via a salvage pathway.Citation48 The flexibility in controlling the fucosylation by simple addition of L-fucose during the manufacturing process offers a great advantage for the RMD system in the generation of fucosylated, afucosylated and even partially fucosylated versions of the same antibody, which can be used for treatment of different therapeutic indications depending on the mode of action.
Table 4b. Difference in EC50 in cell-based potency assay due to reversal of afucosylation by addition of fucose in cell culture medium for RMD expressing mAbA stable cell lines.
Intracellular localization and detection of RMD in secreted mAb and purified IgG samples
The overexpression of RMD in manufacturing host cell lines raises an immunogenicity risk if this protein contributes to the host cell protein load in a mAb bioprocess or contaminates the final product. To address this possibility, we examined the subcellular localization of RMD by immunocytochemistry (). Confocal microscopy was performed on fixed cells co-expressing RMD and IgG, stained simultaneously with allophycocyanin (APC)-labeled anti-FLAG antibody, Alexa-488 labeled antibody against the plasma membrane marker protein alpha 1 sodium potassium ATPase (Atp1a1), and nuclear staining with 4',6-diamidino-2-phenylindole (DAPI). This revealed that RMD is intracellular, mostly localized adjacent to the plasma membrane, but does not overlap with the plasma membrane marker protein in both GS-IRES-RMD and LC-IRES-RMD primary isolates expressing mAb A (). However, subcellular localization of RMD does not confirm whether it secretes with the IgG products. To assess whether RMD is secreted and is co-purified with IgG, we examined cell lysates, crude cell supernatants and purified IgG using high-resolution MS, as well as western blot analyses. An LC-MS/MS analysis of cell lysates from RMD-expressing cells had identified ten peptides derived from RMD with resolution of less than 2.5 parts per million (ppm) (Supplemental Fig. 4, ). Precursors of these peptides were selected during a targeted MS analysis in cell supernatants and purified IgG samples. Our results showed no detectable RMD-derived peptides. While this data does not eliminate the possibility of RMD secretion and purification, it was below the sensitivity of this approach (). RMD was detected by western blot analysis in cell supernatants harvested from LC-IRES-RMD#17 on day eight of a fed-batch fermentation while at high cell viability (97% viable) (C). 50 µg cell lysates were loaded per lane for GS-IRES-RMD#6 and -RMD#20, whereas 5 µg cell lysates were loaded for LC-IRES-RMD#17, 5 µg cell supernatants and purified IgG were analyzed by SDS-PAGE and western blot (C, upper panel). Indeed, RMD was more abundant in LC-IRES-RMD#17 cell supernatant compared to GS-IRES-RMD#6. However, RMD was undetectable by MS and western blot analyses in purified IgG from both GS-IRES-RMD#6 and LC-IRES-RMD#17.These findings suggest that the potential for contamination of purified IgG product with RMD protein is minimal, limiting the risk of immunogenicity against Pseudomonas RMD on in vivo administration.
Figure 6. Intracellular localization and detection of RMD in secreted mAb. A. Localization of recombinant RMD in primary isolate LC-IRES-RMD#17 by confocal microscopy. B. Targeted detection of RMD peptides using mass spectrometry using multiple reaction monitoring. Three distinct peptides unique to RMD were monitored in whole cell lysates, in supernatants of RMD expressing cells and in purified IgG samples. Each peptide was analyzed in isolation to examine the distribution of ions detected (Library). Library indicates mass spectral library of peptides; y ions represent the fragment ions; area is the sum of y ions. The dotP value represents dot product value, which is the similarity extent between targeted spectrum and the library spectrum in the Skyline software. C. Western blot analysis of cell lysates, cell supernatants and purified IgG from GS-IRES-RMD#6, LC-IRES-RMD#17 and -RMD#20 (upper panel). The bottom panel represents a blot used as a loading control where 10 µg cell lysates and 1 µg cell supernatants and purified IgGs were probed with HRP conjugated HC and LC antibodies.
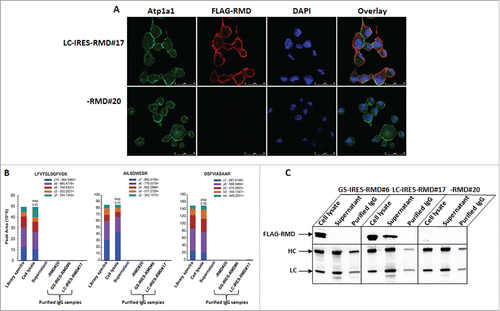
Table 5. List of peptides identified from RMD protein in cell lysates at high confidence peptide level cutoff (1%FDRFootnote1).
Discussion
Antibody glycosylation plays an important role in eliciting activation of ADCC through its interaction with the Fc receptors FcγRIIIa of leukocytes.Citation14,Citation17 This effector function is essential for antibodies used for the treatment of infectious diseases and cancer.Citation16,Citation17 However, antibodies produced in CHO cells often contain oligosaccharides with an α 1–6 linked fucose that has been shown to decrease ADCC activity through lowered binding activity to the FcγRIIIa receptor. Several strategies have been described that enable the expression of afucosylated antibodies from stable CHO cell lines, but these require mutated cell lines that show poor growth propertiesCitation38 or require multiple cell engineering steps.Citation42 Here, we describe a one-step cell line development method by using a single vector for co-expression of RMD and IgG with a platform CHOK1 suspension host, resulting in cell lines capable of producing high yields of afucosylated mAbs and that are stable and scalable using upstream manufacturing platform processes.
Heterologous expression of bacterial RMD in IgG-expressing cells was shown to be an effective means of reducing IgG fucosylation in CHO cells.Citation42 Co-expression of RMD and IgG can be achieved in several ways, including introducing the RMD expression cassette into an IgG-expressing cell lineCitation42 or engineering a CHO host to stably express RMD that would act as the starting point for the introduction of an IgG expression vector.Citation43 Sequential rounds of cell line engineering are time-consuming, so in a third scenario, the RMD and the IgG genes could be expressed simultaneously by introducing independent expression plasmids utilizing different selection markers. However, all three approaches require at least two different selection markers and selection steps, which increase the risk of long-term expression instability upon withdrawal of the selection markers during scale up for manufacturing or increased cost of production by introducing multiple selection reagents during manufacturing.
In an effort to alleviate these issues, we examined methods for the co-expression of IgG and RMD on a single plasmid by exploiting the IRES element that allows mRNA translation initiation in an end-independent manner. Bicistronic RMD vectors were designed with RMD linked via an IRES to either LC or GS, allowing transcription to be driven from a single promoter.Citation49 IRES-dependent genes can show reduced expression levels relative to the upstream gene (expression levels vary from 6 to 100% of the upstream gene) depending on the cell type and genes of interest.Citation50 An alternative approach would be to harness the use of the “ribosomal skipping” mechanism of the viral T2A peptide for an in-frame fusion of multiple open reading frames.51 In our experiments, a wide range of RMD expression was observed using the different expression cassettes tested. Indeed, in the GS-IRES-RMD constructs, expression from the SV40 promoter results in 5-10-fold lower expression of RMD, both at the mRNA and protein levels, than the LC-IRES-RMD construct that used the CMV promoter. However, the levels of IgG afucosylation from these constructs were comparable. These data suggest that minimal expression of RMD is sufficient to deflect the de novo fucosylation pathway ( and ) and produce afucosylated mAbs.
Higher levels of RMD expression were more prevalent in cell lines with the LC-IRES-RMD construct, but we observed a reduction in the number of colonies having detectable titers in the pre-screen. The reduced titers from the LC-IRES-RMD construct screens led us to speculate that LC expression may be reduced, causing an imbalance of HC and LC biosynthesis, with the subsequent excess accumulation of intracellular HC causing cellular toxicity.Citation52 Multiple transient transfection experiments showed limited intracellular LC expression, resulting in undetectable IgG titer while using the LC-IRES-RMD expression cassette (Supplemental Fig. 5). This supports the occurrence of low titer and low expresser frequency in colonies transfected with LC-IRES-RMD for stable IgG expression. Under the influence of the CMV promoter, RMD was highly abundant in the LC-IRES-RMD primary isolates, but no detrimental impact on growth was observed in these cells. Example cell performance data from GS-IRES-RMD#6, LC-IRES-RMD#17 and -RMD#20 cell lines are shown in Supplemental Fig. 6. The cell lines with the GS-linked RMD construct showed overall higher expression titers than those with the LC-linked RMD construct or co-transfected with the two expression plasmids (D).
In summary, we found the GS-IRES-RMD to be an effective vector format that offered high levels of IgG expression and an optimal degree of afucosylation sufficient for maximal ADCC activity. This proof-of-concept study prompted us to apply the GS-IRES-RMD format in live cell line development campaigns for human therapeutic mAbs. The IgG productivity reached or exceeded 6 g/L and showed stable expression over 60 generations in culture while maintaining consistent product quality with undetectable fucosylation in secreted IgG in milliliter scale bioreactors (ambr15™) or even in 3 L bioreactor scale (-C).
Additionally, we demonstrated that the afucosylation can be altered by addition of low amounts of L-fucose to the culture medium during manufacturing, which can be an advantage when the same IgG molecule can be manufactured in fucosylated and afucosylated formats, which is useful for different therapeutic indications requiring diverse effector functions (). Hence, co-expressing RMD with IgG offers greater flexibility over the conventional FUT8 knockout host cells for generating afucosylated mAbs. Furthermore, we demonstrated by confocal microscopy and high-resolution mass spectrometry that the recombinant RMD expressed in CHO cells is intracellular and localized adjacent to cell membranes. If present at high levels, RMD may be secreted along with IgG, but it is not co-purified with the mAb ( and ). Co-expression of the bacterial RMD enzyme in CHO cell lines for therapeutic afucosylated mAb production is thus unlikely to pose a threat to patients by provoking immunogenicity to the RMD proteins.
Materials and methods
Plasmid constructions for co-expression of RMD with mAb A
Three plasmids were constructed for the co-expression of IgG and RMD: 1) A PCR product encoding the Pseudomonas RMD gene was cloned 3′ of a human CMV promoter in a proprietary vector containing a puromycin selection marker by Gibson assembly (pCLD-RMD). The IgG was expressed from a separate vector containing both HC and LC genes driven by the human CMV promoter, along with the GS selectable marker (pCLD-IgG). 2) A GS-IRES-RMD cassette was constructed using overlapping PCR to link GS to RMD with the EMCV-R IRES from pIRES (Takara-Clontech, Mountain View, CA) and then cloned into pCLD-IgG to generate pCLD-IgG GS-IRES-RMD.3) An IRES-RMD cassette was cloned downstream of the IgG LC of pCLD-IgG to generate pCLD-IgG LC-IRES-RMD.
Cell culture and isolation of IgG Expressing primary isolates and clones
IgG-expressing stable primary isolates were generated by Amaxa nucleofection (Lonza, Walkersville, MD) of CHO cells with the IgG expression plasmids described above. Transfected cells were selected in 96-well plates in CD CHO medium (ThermoFisher Scientific, #10743029) supplemented with methionine sulfoximine (Sigma-Aldrich, #M5379) for cells containing pCLD-IgG LC-IRES-RMD or pCLD-IgG HC-IRES-RMD, or MSX and 6 µg/mL puromycin (ThermoFisher Scientific, #1113802) for cells containing the combination of pCLD-IgG and pCLD-RMD. IgG-expressing cell lines were expanded in 384-well plates, 96-deep-well plates and 24-deep-well plates by using Hamilton STAR liquid handling system (Hamilton). Single cell cloning was performed with FACS using a BD Influx cell sorter (Becton, Dickinson and Company). IgG-expressing clones were screened in 96-deep-well plates using a Hamilton automation system. MAb- and RMD-expressing stable cells were evaluated for IgG productivity in a14-day fed-batch assay using MedImmune proprietary production medium and feed regimens. During phenotypic stability studies, cell lines were maintained in E125 flasks and sub-cultured every 3 or 4 days with a viable cell seeding density of 0.3-0.5 × 106 cells/mL in 30 mL volume in a shaking incubator, Multitron (ATR Biotech) at 37°C in the presence of 6% CO2, with 140 RPM and 80% relative humidity. Phenotypic stability studies were carried out in high throughput milliliter scale bioreactor, automated ambr15™ workstations using single-use presterilized Sparge-less ambr15™ micro bioreactors, (TAP Biosystems). Platform bench-scale bioreactor studies were carried out in Applikon 3 L dished-bottom glass bioreactor vessels (2 L working volume) (Chemglass Life Science) for 14-days. IgG titers were quantified by biolayer interferometry using protein A functionalized sensors (Pall ForteBio).
Cell-surface staining of α-linked fucose residues by LCA and analysis by flow cytometry
Cells expressing afucosylated mAbs were identified by surface staining using FITC-conjugated LCA, a lectin that has been shown to bind α-fucose linked residuesCitation34,Citation42 (Vector Laboratories, # FL-1041). Stained cells were analyzed using an LSRII flow cytometer (BD Biosciences) and data processing was performed with FlowJo software (Tree Star Inc). Negatively-stained cells lacking the core fucose moiety represented the RMD-positive population and were carried forward for further analysis.
Purification of IgG
IgG was purified using rProtein A Sepharose Fast Flow resin (GE Healthcare) in a batch mode. rProtein A Sepharose Fast Flow resin was equilibrated using 1X phosphate-buffered saline (PBS), pH 7.2 (ThermoFisher Scientific, #20012050). 50% slurry of resins was incubated with IgG containing conditioned medium derived from GS-IRES-RMD, LC-IRES-RMD or -RMD cell lines for 30 minutes at room temperature followed by elution in 0.1 M glycine, pH 2.8 with a rapid transfer in neutralizing buffer containing 1 M Tris, pH 8.0. The purified mAbs were dialyzed in 1X PBS, pH 7.2 and quantified by Nanodrop A280 using IgG reference.
Western blot analysis
Cells were lysed in mammalian protein extraction reagent, M-PER lysis buffer (Thermo Fisher Scientific, #78501) containing Halt Protease inhibitor (Thermo Fisher Scientific, #78420B,) following the manufacturers’ recommendations. Cell extracts were resolved by SDS-PAGE and transferred to a nitrocellulose membrane using a Trans-Blot Turbo transfer system (Bio-Rad). Membranes were probed with rabbit anti-GS (Abcam, #ab48973) or anti-mouse beta actin (Abcam, #ab8227) antibody which was used as a loading control. Antigen-specific bands were detected by incubation of membranes using horseradish peroxidase (HRP)-conjugated anti-rabbit antibodies followed by incubation with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, #34079) and image analysis with an ImageQuant LAS4000 (GE Heathcare Bio-Sciences). For detection of α-fucose residues in secreted IgG, 2 µg of purified IgG samples were resolved by SDS-PAGE. Biotinylated LCA (Vector Laboratories, #B-1045) was used for binding to the α-fucose residues and visualized using Streptavidin-HRP (GE Healthcare, #RPN1231-100UL). HRP-conjugated anti-human IgG (H+L) was used for detection of HC and LC. FLAG-RMD was detected using HRP-conjugated anti-FLAG antibody (Sigma-Aldrich, #A8592) from crude cell lysates. Due to the differences in expression levels between constructs, 50 µg of crude cell lysate was used for the detection of RMD in GS-IRES-RMD samples whereas 5 µg was used in LC-IRES-RMD samples.
RTPCR analysis and detection of RMD
Total RNA was isolated using an RNeasy plus mini kit (Qiagen, #74134), and reverse transcription (RT) was performed using TaqMan reverse transcription reagents (Thermo Fisher Scientific, #N8080234) with 1 μg RNA according to the manufacturer's protocol. mRNA expression levels were measured using a 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific). The probes and primers for RMD and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were generated by assays-by-design (Applied Biosystems). The probes contained a 5' end 6-carboxy-fluoresceinphosphoramidite (FAM dye) label and a 3' end non-fluorescent quencher. qPCR was performed by an initial denaturation at 95°C for 20 seconds, followed by 40 cycles of 95°C for 1 second and 60°C for 20 seconds. Expression levels of RMD were normalized to endogenous CHO-GAPDH.
HTRF-based CD16a cellular binding assay
The HTRF-based CD16 binding assay was performed using an assay kit from (Cisbio).Citation53 Serial dilutions (1000 µg/ml to 0.7 µg/ml) of purified IgG were incubated at room temperature in 384-well plates with HEK293 cells expressing the CD16a receptor pre-labeled with Terbium donor dye (Tb) in the presence of human IgG labeled with d2 acceptor (IgG-d2) diluted in Tag-lite working buffer. Fluorescence emission was read at 620 nm for the fluorescent donor (Tb) and 665 nm for the fluorescent acceptor (d2) on an EnVision 2104 Multilabel plate reader (Perkin Elmer). The signals were normalized and deltaF/deltaFmax was calculated from the ratio of A665 and A620. All experiments were performed in duplicate and calculations of EC50 were determined using Graph Pad Prism 5.
Localization of RMD and confocal imaging
Actively growing cells were fixed on a coverslip treated with poly-L-lysine (Sigma #P4707) in a 24-well plate with a freshly prepared mixture of methanol and acetone and incubated simultaneously with APC-conjugated anti-FLAG M2 antibody (abcam ab72569) and Alexa-488 labeled anti-alpha 1 sodium potassium ATPase antibody (abcam#ab197496) at room temperature followed by washing with 1x TBS-T (Tween 0.1%). Cells were mounted on a slide in presence of DAPI containing anti-fade mounting agent ProLong Gold Antifade Mountant (ThermoFisher#P36930), then visualized and imaged by confocal microscopy using a Leica SP5 (Leica Microsystems Inc.).
Glycan analysis
N-linked glycans were released using PNGase F (Promega Corporation, V4831) and labeled with aminobenzamide (2-AB). The 2-AB labeled oligosaccharides were cleaned with a HILIC SPE cartridge, eluted into water. Samples were analyzed by UPLC using an Acquity UPLC® BEH Glycan column (Waters), and FLR detection to quantitate the glycoforms present. The peak identification was done by comparison with other know IgG oligosaccharide profiles. 2-AB labeled glycan was further treated with fucosidase (Promega Corporation) to confirm all fucosylated variants.
Intact mass analysis (Reduced)
Quadrupole time-of-flight (Q-TOF) mass spectrometry was used to confirm the glycoforms on the heavy chain based on the accurate measurement of molecular mass. Samples were diluted in 50 mM Tris buffer pH7.8 to 1 mg/ml and reduced by adding dithiothreitol and incubated at 37°C for 30 min. The reduced samples were analyzed by LC/MS using Q-TOF mass spectrometer in conjunction with a UPLC/UV system by reversed-phase chromatography using a BEH C4 column (1.8 μm, 2.1 × 100 mm) in 0.01% trifluoroacetic acid /0.1% formic acid mobile phases with a linear gradient of acetonitrile. Mass spectrometric data were collected with a m/z range of 800–4500. The measured mass of reduced heavy chain was obtained through the spectral mass deconvolution using a maximum entropy deconvolution software package. Glycoforms were identified using the deconvoluted mass compared to the theoretical mass based on the amino acid composition. Relative abundance of each glycoforms were calculated by peak heights of the deconvoluted mass. To confirm the PNGase F cleavage efficiency, the deglycosylated molecule was also analyzed by intact mass to quantify the percentage of deglycosylation.
Detection of RMD in Final IgG product by MS
Identification and quantitation of RMD was carried out using targeted and untargeted tandem MS approaches. Cell lysates containing recombinant RMD were used for the selection of proteotypic peptides.Citation54 Cell lysate (5 µg) was reduced/alkylated and processed through trifluoroethanol-based protocol.Citation55 Peptide samples were analyzed on an Orbitrap Fusion Tribrid™ mass spectrometer with nanoflow liquid chromatography (Dionex RSLCnano™). Initially, data-dependent acquisition (DDA) mode was used to identify the RMD. MS data were searched using Mascot software (v2.5.1) against a database containing all Cricetulus griseus proteins, human IgG HC and LC, and the Pseudomonas aeruginosa RMD sequence.
RMD peptides used for targeted quantification were selected by importing high resolution fragmentation data from tryptic/Lys C peptides of RMD into Skyline software, v3.5. A spectral library was generated from RMD. Tryptic/Lys C peptides were selected for protein quantification. Three peptide precursors of ten detected peptides were included in targeted analysis on Orbitrap Fusion Tribrid® MS.
Abbreviations
| ADCC | = | antibody-dependent cell-mediated cytotoxicity |
| CDC | = | complement-dependent cytotoxicity |
| CHO | = | Chinese hamster ovary |
| IRES | = | internal ribosome entry site |
| LCA | = | Lens culinaris agglutinin |
| MSX | = | methionine sulfoximine |
| RMD | = | GDP-6-deoxy-D-lyxo-4-hexulose reductase |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
supplemenatry__material.zip
Download Zip (5.6 MB)Acknowledgments
We thank Jonathan Boyds and Charles Brown for helping with confocal imaging in RMD localization studies, Nathaniel Macapagal for help with IgG purification, Daphne Fernandes and Thomas Albanetti for long-term stability studies, Aimee Sandjong and Jack Gengcheng Yang for glycan analyses, Weichen Xu for data review of intact mass analyses and Dr. Christopher Sellick for critical review of the manuscript.
References
- Kim JY, Kim YG, Lee GM. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol. 2012;93(3):917–30. Epub 2011/12/14. doi: 10.1007/s00253-011-3758-5. PubMed PMID: 22159888.
- Li F, Vijayasankaran N, Shen A, Kiss R, Amanullah A. Cell culture processes for monoclonal antibody production. MAbs. 2010;2(5):466–77. doi: 10.4161/mabs.2.5.12720. PubMed PMID: 20622510; PubMed Central PMCID: PMCPMC2958569.
- Ghaderi D, Zhang M, Hurtado-Ziola N, Varki A. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnology & genetic engineering reviews. 2012;28:147–75. Epub 2012/05/24. PubMed PMID: 22616486.
- Sola RJ, Griebenow K. Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs: clinical immunotherapeutics, biopharmaceuticals and gene therapy. 2010;24(1):9–21. Epub 2010/01/09. doi: 10.2165/11530550-000000000-00000. PubMed PMID: 20055529; PubMed Central PMCID: PMCPMC2805475.
- Muthana SM, Campbell CT, Gildersleeve JC. Modifications of glycans: biological significance and therapeutic opportunities. ACS chemical biology. 2012;7(1):31–43. Epub 2011/12/27. doi: 10.1021/cb2004466. PubMed PMID: 22195988; PubMed Central PMCID: PMCPMC3262866.
- Higel F, Seidl A, Sorgel F, Friess W. N-glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins. European journal of pharmaceutics and biopharmaceutics: official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik eV. 2016;100:94–100. Epub 2016/01/18. doi: 10.1016/j.ejpb.2016.01.005. PubMed PMID: 26775146
- Reusch D, Tejada ML. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology. 2015;25(12):1325–34. doi: 10.1093/glycob/cwv065. PubMed PMID: 26263923; PubMed Central PMCID: PMCPMC4634315.
- Huang L, Biolsi S, Bales KR, Kuchibhotla U. Impact of variable domain glycosylation on antibody clearance: an LC/MS characterization. Analytical biochemistry. 2006;349(2):197–207. Epub 2005/12/20. doi: 10.1016/j.ab.2005.11.012. PubMed PMID: 16360109.
- Weikert S, Papac D, Briggs J, Cowfer D, Tom S, Gawlitzek M, Lofgren J, Mehta S, Chisholm V, Modi N, et al. Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nature biotechnology. 1999;17(11):1116–21. Epub 1999/11/05. doi: 10.1038/15104. PubMed PMID: 10545921.
- Raju TS. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Current opinion in immunology. 2008;20(4):471–8. Epub 2008/07/09. doi: 10.1016/j.coi.2008.06.007. PubMed PMID: 18606225.
- Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nature reviews Drug discovery. 2009;8(3):226–34. Epub 2009/02/28. doi: 10.1038/nrd2804. PubMed PMID: 19247305.
- Lu J, Chu J, Zou Z, Hamacher NB, Rixon MW, Sun PD. Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(3):833–8. Epub 2015/01/07. doi: 10.1073/pnas.1418812112. PubMed PMID: 25561553; PubMed Central PMCID: PMCPMC4311811.
- Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19(9):936–49. Epub 2009/06/06. doi: 10.1093/glycob/cwp079. PubMed PMID: 19494347.
- Abes R, Teillaud JL. Impact of Glycosylation on Effector Functions of Therapeutic IgG. Pharmaceuticals (Basel, Switzerland). 2010;3(1):146–57. Epub 2010/01/12. doi: 10.3390/ph3010146. PubMed PMID: 27713246; PubMed Central PMCID: PMCPMC3991024.
- Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. The Journal of biological chemistry. 2002;277(30):26733–40. Epub 2002/05/03. doi: 10.1074/jbc.M202069200. PubMed PMID: 11986321.
- Santoli D, Koprowski H. Mechanisms of activation of human natural killer cells against tumor and virus-infected cells. Immunol Rev. 1979;44:125–63. doi: 10.1111/j.1600-065X.1979.tb00269.x. PubMed PMID: 153888.
- Clark MR. IgG effector mechanisms. Chem Immunol. 1997;65:88–110. doi: 10.1159/000319350. PubMed PMID: 9018874.
- Perussia B, Loza MJ. Assays for antibody-dependent cell-mediated cytotoxicity (ADCC) and reverse ADCC (redirected cytotoxicity) in human natural killer cells. Methods in molecular biology (Clifton, NJ). 2000;121:179–92. doi: 10.1385/1-59259-044-6:179. PubMed PMID: 10818726.
- Trapani JA, Smyth MJ. Functional significance of the perforin/granzyme cell death pathway. Nature reviews Immunology. 2002;2(10):735–47. Epub 2002/10/03. doi: 10.1038/nri911. PubMed PMID: 12360212.
- Yamane-Ohnuki N, Satoh M. Production of therapeutic antibodies with controlled fucosylation. MAbs. 2009;1(3):230–6. Epub 2010/01/13. doi: 10.4161/mabs.1.3.8328. PubMed PMID: 20065644; PubMed Central PMCID: PMCPMC2726589.
- Carter PJ. Potent antibody therapeutics by design. Nature reviews Immunology. 2006;6(5):343–57. Epub 2006/04/20. doi: 10.1038/nri1837. doi: 10.1038/nri1837. PubMed PMID: 16622479.
- Davies J, Jiang L, Pan LZ, LaBarre MJ, Anderson D, Reff M. Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: Expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FC gamma RIII. Biotechnol Bioeng. 2001;74(4):288–94. doi: 10.1002/bit.1119. PubMed PMID: 11410853.
- Yu M, Brown D, Reed C, Chung S, Lutman J, Stefanich E, Wong A, Stephan JP, Bayer R. Production, characterization, and pharmacokinetic properties of antibodies with N-linked mannose-5 glycans. mAbs. 2012;4(4):475–87. Epub 2012/06/16. doi: 10.4161/mabs.20737. PubMed PMID: 22699308; PubMed Central PMCID: PMCPMC3499342.
- Boyd PN, Lines AC, Patel AK. The effect of the removal of sialic acid, galactose and total carbohydrate on the functional activity of Campath-1H. Mol Immunol. 1995;32(17-18):1311–8. doi: 10.1016/0161-5890(95)00118-2. PubMed PMID: 8643100.
- Tsuchiya N, Endo T, Matsuta K, Yoshinoya S, Aikawa T, Kosuge E, Takeuchi F, Miyamoto T, Kobata A. Effects of galactose depletion from oligosaccharide chains on immunological activities of human IgG. J Rheumatol. 1989;16(3):285–90. PubMed PMID: 2498512.
- Ritamo I, Cloutier M, Valmu L, Neron S, Rabina J. Comparison of the glycosylation of in vitro generated polyclonal human IgG and therapeutic immunoglobulins. Mol Immunol. 2014;57(2):255–62. doi: 10.1016/j.molimm.2013.10.005. PubMed PMID: 24184880.
- Jiang XR, Song A, Bergelson S, Arroll T, Parekh B, May K, Chung S, Strouse R, Mire-Sluis A, Schenerman M. Advances in the assessment and control of the effector functions of therapeutic antibodies. Nature reviews Drug discovery. 2011;10(2):101–11. Epub 2011/02/02. doi: 10.1038/nrd3365. PubMed PMID: 21283105.
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. The Journal of biological chemistry. 2003;278(5):3466–73. Epub 2002/11/13. doi: 10.1074/jbc.M210665200. PubMed PMID: 12427744.
- Niwa R, Hatanaka S, Shoji-Hosaka E, Sakurada M, Kobayashi Y, Uehara A, Yokoi H, Nakamura K, Shitara K, et al. Enhancement of the antibody-dependent cellular cytotoxicity of low-fucose IgG1 Is independent of FcgammaRIIIa functional polymorphism. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(18 Pt 1):6248–55. Epub 2004/09/28. doi: 10.1158/1078-0432.ccr-04-0850. PubMed PMID: 15448014.
- Niwa R, Shoji-Hosaka E, Sakurada M, Shinkawa T, Uchida K, Nakamura K, Matsushima K, Ueda R, Hanai N, Shitara K. Defucosylated chimeric anti-CC chemokine receptor 4 IgG1 with enhanced antibody-dependent cellular cytotoxicity shows potent therapeutic activity to T-cell leukemia and lymphoma. Cancer research. 2004;64(6):2127–33. Epub 2004/03/18. doi: 10.1158/0008-5472.CAN-03-2068. PubMed PMID: 15026353.
- Niwa R, Natsume A, Uehara A, Wakitani M, Iida S, Uchida K, Satoh M, Shitara K.. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. Journal of immunological methods. 2005;306(1-2):151–60. Epub 2005/10/13. doi: 10.1016/j.jim.2005.08.009. PubMed PMID: 16219319.
- Niwa R, Sakurada M, Kobayashi Y, Uehara A, Matsushima K, Ueda R, Nakamura K, Shitara K. Enhanced natural killer cell binding and activation by low-fucose IgG1 antibody results in potent antibody-dependent cellular cytotoxicity induction at lower antigen density. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(6):2327–36. Epub 2005/03/25. doi: 10.1158/1078-0432.ccr-04-2263. PubMed PMID: 15788684.
- Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, et al. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology. 2007;17(1):104–18. Epub 2006/10/03. doi: 10.1093/glycob/cwl057. PubMed PMID: 17012310.
- Kanda Y, Yamane-Ohnuki N, Sakai N, Yamano K, Nakano R, Inoue M, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, et al. Comparison of cell lines for stable production of fucose-negative antibodies with enhanced ADCC. Biotechnol Bioeng. 2006;94(4):680–8. Epub 2006/04/13. doi: 10.1002/bit.20880. PubMed PMID: 16609957.
- Suzuki E, Niwa R, Saji S, Muta M, Hirose M, Iida S, Shiotsu Y, Satoh M, Shitara K, Kondo M, et al. A nonfucosylated anti-HER2 antibody augments antibody-dependent cellular cytotoxicity in breast cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(6):1875–82. Epub 2007/03/17. doi: 10.1158/1078-0432.ccr-06-1335. PubMed PMID: 17363544.
- Alves CS, Prajapati S. Optimizing Chinese hamster ovary cell line development via targeted control of N-glycosylation. Pharmaceutical Bioprocessing. 2015;3(7):443–61. doi:10.4155/pbp.15.25.
- Kunert R, Reinhart D. Advances in recombinant antibody manufacturing. Appl Microbiol Biotechnol. 2016;100(8):3451–61. Epub 2016/03/05. doi: 10.1007/s00253-016-7388-9. PubMed PMID: 26936774; PubMed Central PMCID: PMCPMC4803805.
- Yamane-Ohnuki N, Kinoshita S, Inoue-Urakubo M, Kusunoki M, Iida S, Nakano R, Wakitani M, Niwa R, Sakurada M, Uchida K, et al. Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnology and bioengineering. 2004;87(5):614–22. doi: 10.1002/bit.20151. PubMed PMID: 15352059.
- Mori K, Kuni-Kamochi R, Yamane-Ohnuki N, Wakitani M, Yamano K, Imai H, Kanda Y, Niwa R, Iida S, Uchida K, et al. Engineering Chinese hamster ovary cells to maximize effector function of produced antibodies using FUT8 siRNA. Biotechnology and bioengineering. 2004;88(7):901–8. Epub 2004/10/30. doi: 10.1002/bit.20326. PubMed PMID: 15515168.
- Kanda Y, Imai-Nishiya H, Kuni-Kamochi R, Mori K, Inoue M, Kitajima-Miyama K, Okazaki A, Iida S, Shitara K, Satoh M. Establishment of a GDP-mannose 4,6-dehydratase (GMD) knockout host cell line: a new strategy for generating completely non-fucosylated recombinant therapeutics. Journal of biotechnology. 2007;130(3):300–10. Epub 2007/06/15. doi: 10.1016/j.jbiotec.2007.04.025. PubMed PMID: 17559959.
- Ho DT PJ, Ho DE, Nunez B, Bang J and Ni JHT. Fucosylation of a Therapeutic Antibody Effects on Antibody-Dependent, Cell-Mediated Cytotoxicity (ADCC) Potency and Efficacy. BioProcess International. 2016;14(4):30–8.
- von Horsten HH, Ogorek C, Blanchard V, Demmler C, Giese C, Winkler K, Kaup M, Berger M, Jordan I, Sandig V. Production of non-fucosylated antibodies by co-expression of heterologous GDP-6-deoxy-D-lyxo-4-hexulose reductase. Glycobiology. 2010;20(12):1607–18. Epub 2010/07/20. doi: 10.1093/glycob/cwq109. PubMed PMID: 20639190.
- Puklowski A, Wenger T, Schatz S, Koenitzer J, Schaub J, Enenkel B, et al. BI-HEX(®)-GlymaxX(®)cells enable efficient production of next generation biomolecules with enhanced ADCC activity: BMC Proc. 2013;7(Suppl 6):P63. doi:10.1186/1753-6561-7-S6-P63.
- Stewart R, Morrow M, Hammond SA, Mulgrew K, Marcus D, Poon E, Watkins A, Mullins S, Chodorge M, Andrews J, et al. Identification and Characterization of MEDI4736, an Antagonistic Anti-PD-L1 Monoclonal Antibody. Cancer immunology research. 2015;3(9):1052–62. Epub 2015/05/07. doi: 10.1158/2326-6066.cir-14-0191. PubMed PMID: 25943534.
- Li X, Kimberly RP. Targeting the Fc receptor in autoimmune disease. Expert opinion on therapeutic targets. 2014;18(3):335–50. doi: 10.1517/14728222.2014.877891. PubMed PMID: 24521454; PubMed Central PMCID: PMCPMC4019044.
- Chung S, Quarmby V, Gao X, Ying Y, Lin L, Reed C, Fong C, Lau W, Qiu ZJ, Shen A, et al. Quantitative evaluation of fucose reducing effects in a humanized antibody on Fcγ receptor binding and antibody-dependent cell-mediated cytotoxicity activities. MAbs. 2012;4(3):326–40. doi: 10.4161/mabs.19941. PubMed PMID: 22531441; PubMed Central PMCID: PMCPMC3355491.
- Evans K, Albanetti T, Venkat R, Schoner R, Savery J, Miro-Quesada G, Rajan B, Groves C, et al. Assurance of monoclonality in one round of cloning through cell sorting for single cell deposition coupled with high resolution cell imaging. Biotechnology progress. 2015;31(5):1172–8. Epub 2015/07/22. doi: 10.1002/btpr.2145. PubMed PMID: 26195345; PubMed Central PMCID: PMCPMC5054913.
- Niittymaki J, Mattila P, Renkonen R. Differential gene expression of GDP-L-fucose-synthesizing enzymes, GDP-fucose transporter and fucosyltransferase VII. Apmis. 2006;114(7-8):539–48. Epub 2006/08/16. doi: 10.1111/j.1600-0463.2006.apm_461.x. PubMed PMID: 16907860.
- Lai T YY, Ng SK. Advances in Mammalian Cell Line Development Technologies for Recombinant Protein Production. Pharmaceuticals. 2013;6:579–603. doi: 10.3390/ph6050579.
- Mizuguchi H, Xu Z, Ishii-Watabe A, Uchida E, Hayakawa T. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther. 2000;1(4):376–82. Epub 2000/08/10. doi: 10.1006/mthe.2000.0050. PubMed PMID: 10933956.
- Diao F, White BH. A Novel Approach for Directing Transgene Expression in Drosophila: T2A-Gal4 In-Frame Fusion. Genetics. 2012;190(3):1139–44. doi: 10.1534/genetics.111.136291. PubMed PMID: 22209908; PubMed Central PMCID: PMCPMC3296248.
- Hendershot LM, Ting J, Lee AS. Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Molecular and cellular biology. 1988;8(10):4250–6. Epub 1988/10/01. PubMed PMID: 3141786; PubMed Central PMCID: PMCPMC365497.
- Degorce F. HTRF((R)): pioneering technology for high-throughput screening. Expert opinion on drug discovery. 2006;1(7):753–64. Epub 2006/12/01. doi: 10.1517/17460441.1.7.753. PubMed PMID: 23495998.
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25(1):125–31. doi: 10.1038/nbt1275. PubMed PMID: 17195840.
- Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, Camp DG 2nd, Khan AH, Sforza DM, Pallavicini M, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4(6):2397–403. Epub 2005/12/13. doi: 10.1021/pr050160f. PubMed PMID: 16335993; PubMed Central PMCID: PMCPMC1781925.

