Abstract
Background
Short tandem repeats (STR) are highly polymorphic DNA markers utilised in forensic personal identification and human population genetic research. Guizhou Tujia is one of the ancient minority groups in southwest China, however, the population has not been studied using the highly discriminating 23 STR Huaxia Platinum Kit.
Aim
To obtain genetic data from 23 autosomal STRs in Guizhou Tujia and examine the population’s relationship with others.
Subjects and methods
A total of 480 individuals from the Guizhou Tujia population were analysed using 23 STR loci of Huaxia Platinum Kit. Allele frequencies and forensic parameters were estimated. Population genetic relationships were calculated by Nei’s genetic distances and visualised using a variety of biostatistical methods.
Results
A total of 264 alleles were found, with allelic frequencies ranging from 0.0010 to 0.5104. The combined discrimination power (CDP) and the combined probability of paternity (CPE) of 23 STR loci were 0.9999999999999999999999999996 and 0.999999999710422, respectively. Guizhou Tujia showed closer genetic relationships with Hubei Tujia, Guizhou Gelao, and Guizhou Miao than with other populations.
Conclusion
We first obtained the population genetic data of Guizhou Tujia using the 23 STR system and demonstrated its value in forensic applications. Comprehensive population comparisons showed an evident genetic affinity pattern between populations that are geographically, ethnically and linguistically related.
Background
Short tandem repeats (STRs) are repetitive nucleotide sequences ranging from 2 to 6 base pairs widely distributed throughout the human genome (Edwards et al. Citation1991; Ellegren Citation2004; Jobling and Gill Citation2004; Chen et al. Citation2018). Due to their high level of polymorphic information, STR markers have become the gold standard in forensic DNA analysis, especially in human identification and paternity testing, surpassing other DNA markers such as single nucleotide polymorphisms (SNPs) (Zhang Citation2015; Yao and Wang Citation2016; He et al. Citation2018; Wu et al. Citation2020). In recent years, new commercial multiple STR detection systems containing more autosomal STRs have been developed to further improve their discriminating ability with the increasing growth of forensic DNA databases. Additionally, STRs are frequently used in human population genetics to predict the population genetic structure of geographically and ethno-linguistically diverse subpopulations via genetic affinity analysis (Gao et al. Citation2021; Kumar et al. Citation2021; Tran et al. Citation2021; Wang et al. Citation2021; Chandra et al. Citation2022).
Guizhou province, located in southwest China, is democratically one of the most ethnically diverse provinces in the country, home to 56 ethnic groups, 17 of which are ancient indigenous ethnic minorities. Among these groups are the Tujia, an ancient Chinese ethnic group residing mainly on the border of Guizhou, Hunan, Hubei, and Chongqing. The Guizhou Tujia population accounts for approximately 4.1% of all Tujia people in China, making them the fifth largest ethnic group in Guizhou province (https://en.wikipedia.org/wiki/Guizhou). The ancestry of Tujia can be traced to the ancient Ba people who lived in southwest China during the Xia and the Shang dynasties. Furthermore, their unique language is categorised into the Tibeto-Burman branch of the Sino-Tibetan language family.
Currently, many geographically, ethnically, and linguistically distinct populations in China are being studied using the Huaxia Platinum Kit, which was developed as a highly discriminating system to characterise Chinese population genetics (Wang et al. Citation2015; Ludeman et al. Citation2018). However, there is limited population data available for the Guizhou Tujia, hampering their use in forensic DNA cases and population genetic studies. To address this issue, we genotyped 23 autosomal STRs in 480 Guizhou Tujia individuals using the Huaxia Platinum Kit to provide a more precise population genetic dataset for forensic practice and human population genetics.
Sample
Blood samples were collected using the FTA cards from 480 unrelated healthy Tujia individuals, comprising 254 males and 226 females, in the Tongren district of northeast Guizhou (Figure S1). All volunteers were self-identified indigenous, had no history of immigration or interracial marriage within three generations, and provided informed signed consent before sample collection. The project was approved by the Biomedical Research Ethics Committee of the Affiliated Hospital of Zunyi Medical University (ID: KLLY-2021-110).
Data collection
Genomic DNA was extracted from the blood samples according to the Chelex-100 protocol (Walsh et al. Citation1991). Multiplex PCR was performed using the Huaxia Platinum Kit (Thermo Fisher Scientific) following the manufacturer’s guidelines on the GeneAmp® 9700 PCR system (Thermo Fisher Scientific). PCR products were detected by capillary electrophoresis on an Applied Biosystems® 3500 Genetic Analyser (Thermo Fisher Scientific). Alleles were assigned by comparing sample PCR fragments with allelic ladders provided with the kit using the GeneMapper ID-X V1.4 software (Thermo Fisher Scientific). The negative control (H2O) and positive control (007 DNA) were simultaneously detected in each batch to ensure reliable analytical results and no reagent contamination.
Data management and statistical analysis
Allelic frequencies and forensic descriptive parameters, including the observed heterozygosity (Ho), polymorphism information content (PIC), discrimination power (PD), probability of exclusion (PE), match probability (PM) and typical paternity index (TPI), were calculated using the online software STR Analysis for Forensics (STRAF) (Gouy and Zieger Citation2017). Hardy-Weinberg Equilibrium (HWE) and Linkage Disequilibrium (LD) tests were performed with the Arlequin V3.5 software (Excoffier and Lischer Citation2010).
The total population genetic relationships of Guizhou Tujia and 22 reference populations (17 Chinese groups and 5 non-Chinese groups) selected based on the allele frequency data of 23 overlapping STRs were revealed using biostatistical methods. Pairwise Nei’s genetic distances based on allele frequency distribution were computed using the Phylogeny Inference Packages (genedist package) of the Phylip software V3.695(Ropelewski et al. Citation2010). Based on Nei’s genetic distances, further clear genetic phylogenetic relationships among these 23 populations were illustrated and visualised via heat maps using the Tbtools V1.0 Software (Chen et al. Citation2020). Additionally, principal component analysis (PCA) and multi-dimensional scaling plots (MDS) were conducted using SPSS V26.0(Hansen Citation2005), and the neighbor-joining tree (N-J tree) was constructed using the Mega V7.0 software (Kumar et al. Citation2016).
Results and discussions
The allele frequency distributions and related forensic parameters for the 23 STR loci are summarised in Table S1. In total 264 alleles were screened with allelic frequencies ranging from 0.0010∼0.5104. After Bonferroni correction, each locus was consistent with the HWE (0.05/23 ≈ 0.002174) (Table S1), with no significant LD found in any locus pair (0.05/253 ≈ 0.000198) (Table S2). Ho and PIC varied from 0.9271 to 0.6250, and 0.5666 to 0.9071, respectively. Penta E had the highest genetic polymorphism, while TPOX had the lowest. PD, PE, TPI, and PM ranged from 0.8043 to 0.9837, 0.3220 to 0.8510, 1.3333 to 6.8571, and 0.0163 to 0.1957, respectively. The combined power of discrimination (CPD) and the combined power of exclusion (CPE) of the 23 STR system were 0.9999999999999999999999999996 and 0.999999999710422, respectively. As displayed by the data above, these 23 STRs in the Huaxia Platinum STR Kit exhibited high genetic polymorphism and discriminating power in the investigated Guizhou Tujia minority, demonstrating potential use in forensic cases such as human identity testing and paternity testing. The resulting population genetic data of the Tujia minority can significantly enrich the forensic database of Chinese ethnicities and be used in forensic and anthropological population genetic research.
To obtain a comprehensive and clear understanding of the genetic differences and correlations between Guizhou Tujia and others, we first analysed the genetic relationship based on the allele frequency distribution of the 23 overlapped STRs. A total of 22 reference populations from three language families -the Sino-Tibetan, Altaic and Indo-European- were selected for comparison through various analyses. The 14 populations of the Sino-Tibetan language family were divided into four language branches: 4 Tibeto-Burman (Hubei Tujia [abbreviated as HBT], Sichuan-Liangshan Tibetan [SCT1], Sichuan-Chengdu Tibetan [SCT1], Sichuan Yi [SCY]), 8 Sinitic (Guizhou Han [GZH], Hunan Hui [HNH], Shandong Han [SDH], Shaanxi Han [SHXH], Shanxi Han [SXH], Hainan Han [HANH], Liaoning Han [LNH], Sichuan Han [SCH], 1 Tai-Kadai (Guizhou Gelao [GZG]), and 1 Hmong-Mien (Guizhou Miao [GZM)] (Shen et al. Citation2014; He et al. Citation2018; Wang et al. Citation2018; Yang and Hu Citation2018; Chen, Wu, et al. Citation2019a, Citation2019b ; Liu et al. Citation2019a; Citation2019b; Zhang et al. Citation2019; Li et al. Citation2020; Liu et al. Citation2020; Zhang et al. Citation2020). Meanwhile, 3 Altaic language family populations in this study were all of the Turkic language branches (Xinjiang Kyrgyz [XJK], Xinjiang Kazakh [XJKA], and Xinjiang Uyghur [XJU]) (Jin et al. Citation2017; Chen, Zou, et al. 2019a, Citation2019b; Liu et al. Citation2019a, Citation2019b). The 5 reference populations of the Indo-European language family consisted of US African American [UAA], US Hispanic [UH], White British [WB], Northeast Colombian [NC] and Indian [ID] (Hill et al. Citation2013; Castilloa et al. Citation2019; Krzeminska-Ahmadzai et al. Citation2021; Shrivastava et al. Citation2022).
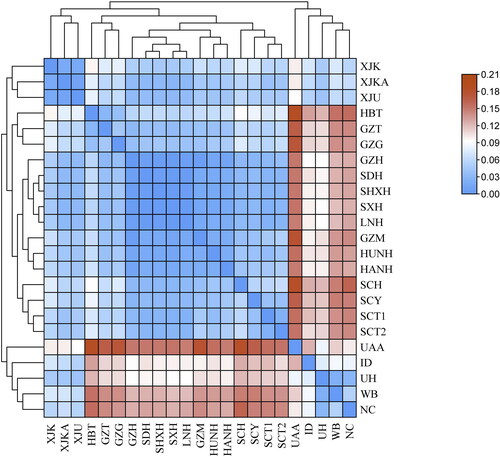
The Nei’s genetic distance among the investigated 23 populations is presented in Table S3 and illustrated in via a heatmap. The closest genetic distances were found between Guizhou Tujia and Hubei Tujia populations (0.0283). However, Guizhou Tujia exhibited the largest genetic distances with the Turkic-speaking Xinjiang Kyrgyz population (0.0765) among the 17 referenced Chinese and African American populations (0.1669) out of the 5 compared non-Chinese populations.
Figure 1. Heatmap of pairwise Nei’s genetic distance between the Guizhou Tujia population and other 22 reference populations. The population names are displayed as in abbreviated form (similar to other figures and tables), and the genetic affinity and genetic differences of the Guizhou Tujia with others are demonstrated using distinct colours.
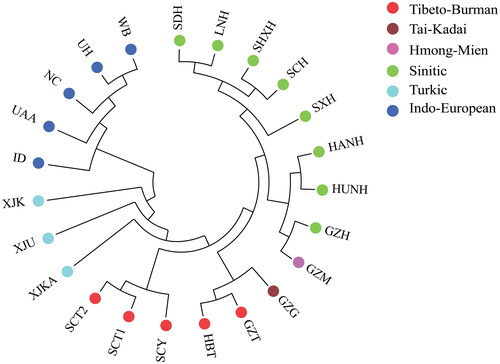
The phylogenetic tree presented in shows that the 23 populations analysed could be divided into two main branches: one containing the five non-Chinese groups (clustered into one branch) and the other including 18 Chinese groups. Within the branch of 18 Chinese groups, several distinct clusters were identified. The Guizhou Tujia population was first grouped with the Hubei Tujia from the adjacent province and then merged with the Tai-Kadai-speaking Guizhou Gelao. All Sinitic-speaking Han groups from different provinces were grouped into one independent cluster. Furthermore, the Guizhou Han and the Hmong-Mien-speaking Guizhou Miao from the same region were also closely gathered. Populations that are both geographically closer and belong to the same language branch, such as the Tibeto-Burman- speaking Sichuan Yi and Sichuan Tibetan populations, as well as the Turkic-speaking Xinjiang Uyghurs, Xinjiang Kyrgyz and Xinjiang Kazakhs, were prominently gathered together. The phylogenetic tree demonstrated typical genetic affinity patterns based on their geographical, ethical, and linguistic origins.
Figure 2. Neighbor-joining tree based on Nei’s distance of the Guizhou Tujia population and 22 reference populations. The language families/branches they belonged to are denoted using different colour circles.
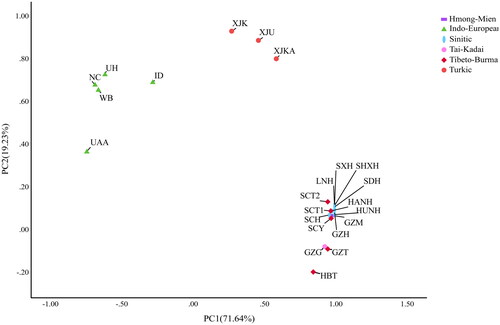
As shown by the PCA analysis (), the first two components extracted a total of 90.87% genetic variance (PC1:71.64%; PC2: 19.23%) and separated the 23 populations into three main distinct clusters: the five Indo-European-speaking populations cluster; the Turkic-speaking populations cluster; and the remaining 15 Sino-Tibetan populations cluster, which belongs to Sinitic, Tibeto-Burman, Tai-Kadai, and Hmong-Mien language branches. As for the third cluster, the Guizhou Tujia and Guizhou Gelao were found to be very close to each other, and the Sinitic Han populations were tightly assembled.
Figure 3. Principal component analysis (PCA) based on Nei’s distance of the Guizhou Tujia population and 22 reference populations. The 23 populations are separated into three distinct main language family clusters, the Sino-Tibetan, the Altaic, and the Indo-European.
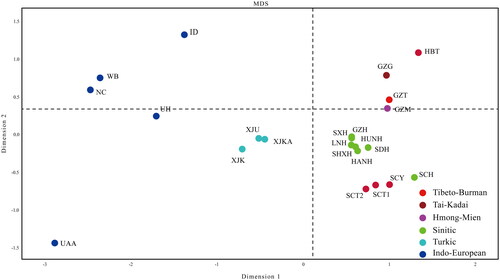
To further illustrate the genetic similarities and differences of the examined populations, a two-dimensional MDS plot was constructed (). We observed that the three Guizhou minorities (Guizhou Tujia, Guizhou Miao, and Guizhou Gelao) and the Hubei Tujia were gathered in the first quadrant. Except for the Sichuan Han, all Han populations from diverse administrative divisions were tightly aggregated in the upper left of the fourth quadrant. The 4 populations in Sichuan Province (Sichuan Han, Sichuan Yi, Sichuan Chengdu Tibetan, and Sichuan Liangshan Tibetan) were grouped in the middle part of the fourth quadrant. Further, the Indo-European-speaking populations were scattered on the left side of the MDS plot, while the Turkic-speaking populations were gathered in the centre.
Figure 4. The Multidimensional Scaling plots (MDS) displayed the genetic relationships between the Guizhou Tujia population and 22 reference populations based on 23 overlapping STRs. Several clusters were formed according to their linguistical/geographical origin.
Comprehensive population genetic analysis using multiple analytical methods revealed an overall consistent genetic affinity pattern. The Guizhou Tujia demonstrated similarity with the Guizhou Miao, Guizhou Gelao and Hubei Tujia populations, which can be attributed to extensive gene exchange due to long-term historical migration and intermarriage between the four populations residing in adjacent geographic areas. Notably, the Guizhou Tujia and Hubei Tujia populations share not only geographic proximity but also a common ancestral ethnic/linguistic background, thereby indicating a higher degree of ancestral components shared between them than other populations. The observed correlations in Guizhou Tujia in this study were similar to previous reports from analyses of other autosomal STRs, X-STRs and Y-STRs, further supporting our results (Chen, Wu, et al. Citation2019; Luo et al. Citation2020; Citation2021). Similar genetic affinity patterns shaped by geographic, ethnic and linguistic factors were also evident in other population groups, such as the aforementioned Sinitic-speaking Chinese Han populations, the Sinitic and Tibeto-Burman speaking populations of Sichuan province, as well as the Turkic-speaking populations of Xinjiang region.
Conclusion
In this study, we employed the Huaxia Platinum Kit for the first time to investigate genetic polymorphism in the Tujia population of Guizhou, and our results indicate that 23 loci could be valuable for human identification and paternity testing. Additionally, our population comparisons by diverse analytical methods revealed an evident genetic affinity pattern between geographically, ethnically and linguistically related populations. The Guizhou Tujia has closer genetic relationships with Hubei Tujia, Guizhou Gelao, and Guizhou Miao than with the other populations. These findings add to the growing body of knowledge on genetic variation and population history in China and provide insights into the complex interplay between genetic, geographic, ethnic, and linguistic factors that shape population diversity.
Supplemental Material
Download Zip (307.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Castilloa A, PicoaA A, GilaA A, Gusmãob L, Vargasa C. 2019. Genetic variation of 23 STR loci in a Northeast Colombian population (department of Santander). Forensic Sci Int. 7(2019):33–35.
- Chandra D, Mishra VC, Raina A, Raina V. 2022. Mutation rate evaluation at 21 autosomal STR loci: paternity testing experience. Leg Med. 58:102080. doi: 10.1016/j.legalmed.2022.102080.
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. 2020. TBtools: an integrative Toolkit developed for interactive analyses of big biological data. Mol Plant. 13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009.
- Chen L, Guo Y, Xiao C, Wu W, Lan Q, Fang Y, Chen J, Zhu B. 2018. Genetic polymorphisms and forensic efficiency of 19 X-chromosomal STR loci for Xinjiang Mongolian population. PeerJ. 6:e5117. doi: 10.7717/peerj.5117.
- Chen P, Wu J, Luo L, Gao H, Wang M, Zou X, Li Y, Chen G, Luo H, Yu L, et al. 2019. Population genetic analysis of modern and ancient DNA variations yields new insights into the formation, genetic structure, and phylogenetic relationship of Northern Han Chinese. Front Genet. 10:1045. doi: 10.3389/fgene.2019.01045.
- Chen P, Zou X, Wang B, Wang M, He G. 2019. Genetic admixture history and forensic characteristics of Turkic-speaking Kyrgyz population via 23 autosomal STRs. Ann Hum Biol. 46(6):498–501. doi: 10.1080/03014460.2019.1676918.
- Edwards A, Civitello A, Hammond HA, Caskey CT. 1991. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet. 49(4):746–756.
- Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 5(6):435–445. eng. doi: 10.1038/nrg1348.
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x.
- Gao H, Yu J, Feng X, Wu X, Luo L, Li X, Liu C, Chen P. 2021. Genetic polymorphism of 23 autosomal STR loci in Han population from Yuncheng, Shanxi Province. Zhong nan da xue xue bao Yi xue ban. J Cent South Univ Med Sci. 46(4):351–360.
- Gouy A, Zieger M. 2017. STRAF-A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci Int Genet. 30:148–151. doi: 10.1016/j.fsigen.2017.07.007.
- Hansen J. 2005. Using SPSS for Windows and Macintosh: analyzing and understanding data. American Statistician. 59(1):113–113. doi: 10.1198/tas.2005.s139.
- He G, Wang M, Liu J, Hou Y, Wang Z. 2018. Forensic features and phylogenetic analyses of Sichuan Han population via 23 autosomal STR loci included in the Huaxia Platinum System. Int J Legal Med. 132(4):1079–1082. doi: 10.1007/s00414-017-1679-2.
- Hill CR, Duewer DL, Kline MC, Coble MD, Butler JM. 2013. U.S. population data for 29 autosomal STR loci. Forensic Sci Int Genet. 7(3):e82–e83. doi: 10.1016/j.fsigen.2012.12.004.
- Jin X, Wei Y, Chen J, Kong T, Mu Y, Guo Y, Dong Q, Xie T, Meng H, Zhang M, et al. 2017. Phylogenic analysis and forensic genetic characterization of Chinese Uyghur group via autosomal multi STR markers. Oncotarget. 8(43):73837–73845. doi: 10.18632/oncotarget.17992.
- Jobling MA, Gill P. 2004. Encoded evidence: DNA in forensic analysis. Nat Rev Genet. 5(10):739–751. doi: 10.1038/nrg1455.
- Krzeminska-Ahmadzai U, Buckley B, Loake T, Nicholson C, Beesley D, Randall C. 2021. Population data for 23 autosomal STR loci in White British population. Leg Med. 50:101863. doi: 10.1016/j.legalmed.2021.101863.
- Kumar A, Kumar R, Kumawat RK, Shrivastava P, Yadav R, Chaubey G. 2021. Population genetic data of 22 autosomal STR loci for the Scheduled Caste population of Rajasthan India. Ann Hum Biol. 48(7–8):598–604. doi: 10.1080/03014460.2021.2009030.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Li L, Zou X, Zhang G, Wang H, Su Y, Wang M, He G. 2020. Population genetic analysis of Shaanxi male Han Chinese population reveals genetic differentiation and homogenization of East Asians. Mol Genet Genomic Med. 8(5):e1209. doi: 10.1002/mgg3.1209.
- Liu Y, Li J, Yue J, Li X, Shi M. 2019a. Genetic polymorphisms and genetic relationship of 24 autosomal STR loci in Gelao and Miao population of Guizhou. Med J Chin PLA. 44(08):682–689.
- Liu Y, Li X, Guo L, Li X, Shi M. 2019b. Genetic polymorphisms and their relationship of 23 autosomal STR loci in Kazakh population of Xinjiang. Basic Clin Med. 39(02):157–164.
- Liu Y, Yue J, Li J, Li X, Shi M. 2020. Genetic polymorphisms of 24 short tandemrepeat loci in Hubei Tujia population. Lab Med Clin. 17(13):1800–1804.
- Ludeman MJ, Zhong C, Mulero JJ, Lagacé RE, Hennessy LK, Short ML, Wang DY. 2018. Developmental validation of GlobalFiler™ PCR amplification kit: a 6-dye multiplex assay designed for amplification of casework samples. Int J Legal Med. 132(6):1555–1573. doi: 10.1007/s00414-018-1817-5.
- Luo L, Gao H, Yao L, Long F, Zhang H, Zhang L, Liu Y, Yu J, Yu L, Chen P. 2020. Genetic diversity, forensic feature, and phylogenetic analysis of Guizhou Tujia population via 19 X-STRs. Mol Genet Genomic Med. 8(11):e1473. doi: 10.1002/mgg3.1473.
- Luo L, Yao L, Chai S, Zhang H, Li M, Yu J, Hu X, Li C, Bian Y, Chen P. 2021. Forensic characteristics and population construction of two major minorities from southwest China revealed by a novel 37 Y-STR loci system. R Soc Open Sci. 8(7):210447. doi: 10.1098/rsos.210447.
- Ropelewski AJ, Nicholas HB, Gonzalez Mendez RR. 2010. MPI-PHYLIP: parallelizing computationally intensive phylogenetic analysis routines for the analysis of large protein families. PLOS One. 5(11):e13999. doi: 10.1371/journal.pone.0013999.
- Shen H, Guo F, Jin P, Yu J, Jia F, Zhao J, Jiang X. 2014. Genetic polymorphisms of 23 STR loci in Han populations in Liaoning. Chin J Forensic Med. 29(03):259–261.
- Shrivastava P, Dixit S, Kumawat RK, Srivastava A. 2022. Efficiency analysis of VersaPlex™ 27PY system in Central Indian population: first report from Indian population. Leg Med. 54:101983. doi: 10.1016/j.legalmed.2021.101983.
- Tran HL, Ta TAM, Nguyen NN, Pham TT, Hoang H, Ha Chu H. 2021. Population genetic data of 22 autosomal STR loci for the Mong people in Vietnam. Leg Med). 48:101825. doi: 10.1016/j.legalmed.2020.101825.
- Walsh PS, Metzger DA, Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 10(4):506–513.
- Wang DY, Gopinath S, Lagacé RE, Norona W, Hennessy LK, Short ML, Mulero JJ. 2015. Developmental validation of the GlobalFiler(®) Express PCR Amplification Kit: a 6-dye multiplex assay for the direct amplification of reference samples. Forensic Sci Int Genet. 19:148–155. doi: 10.1016/j.fsigen.2015.07.013.
- Wang H, Xin C, Cao H. 2021. Forensic features and genetic structure of 20 autosomal STR loci in the Han population of Ningde City, Southeastern China. Ann Hum Biol. 48(1):56–61. doi: 10.1080/03014460.2020.1854344.
- Wang M, Wang Z, He G, Jia Z, Liu J, Hou Y. 2018. Genetic characteristics and phylogenetic analysis of three Chinese ethnic groups using the Huaxia Platinum system. Sci Rep. 8(1):2429. doi: 10.1038/s41598-018-20871-7.
- Wu X, Zheng JL, Lou Y, Wei XH, Wang BJ, Yao J. 2020. Genetic polymorphisms of 20 autosomal STR loci in the Han population of Zhangzhou City, Southeastern China. Leg Med. 46:101726. doi: 10.1016/j.legalmed.2020.101726.
- Yang J, Hu R. 2018. Application of Goldeneye 25A identification system in the HUI population. Forensic SciInt. 2018(01):72–75.
- Yao J, Wang BJ. 2016. Genetic variation of 25 Y-chromosomal and 15 autosomal STR Loci in the Han Chinese Population of Liaoning Province, Northeast China. PLOS One. 11(8):e0160415. doi: 10.1371/journal.pone.0160415.
- Zhang H, Hu X, Liu Z, Luo L, Wang W, Cheng P, Yu L. 2020. Study on OL alleles of 23 STR loci in Guizhou Han population. J Zunyi Med Univ. 43(06):686–691.
- Zhang J, Zhao Z, Li L, Li X, Liu Z. 2019. Validation of HuaxiaTM Platinum PCR amplification Kit and the STR genetic polymorphism of Shandong Population. Forensic SciInt. 2019(06):41–48.
- Zhang L. 2015. Population data for 15 autosomal STR loci in the Bouyei ethnic minority from Guizhou Province, Southwest China. Forensic Sci Int Genet. 17:108–109. doi: 10.1016/j.fsigen.2015.04.006.
