Abstract
Genetic heterogeneity and polymorphism in muscle proteins present in the meat of wild and cultured Labeo rohita and Cirrhinus mrigala was performed using SDS–PAGE. Cultured L. rohita and C. mrigala possessed more proteins than those in the wild ones and were dependent on their weight. Un-weighed pair group method with arithmetic average cluster analysis delineated a 50% linkage distance. Fish weight dissimilarity coefficient ranged from 1.73 to 3.87 in eight clusters at 75% linkage distance. The overall grouping pattern of clusters corresponded well with principal co-ordinate analysis and confirmed overall patterns of genetic variability among these species. These results provide useful guidelines for conservation and characterization of fish genetic resources for mass rearing or marker assisted breeding.
INTRODUCTION
The studies on nutritional status of fish show that fish proteins from marine species are good sources of high-quality proteins and polyunsaturated fatty acids. There is an opportunity to use more fish by-products as protein ingredients for foods, feeds, and industrial application.[Citation1–4 Citation Citation Citation4 Fish is a major muscle protein source and considered to be superior to that of other proteins.[Citation5] A unique feature of fish muscle is its low connective tissue content, which accounts for easy disintegration of fish flesh on heating.[Citation6–8 Citation Citation8 The myofibril proteins, namely, myosin, actin, tropomyosin, and troponin, are involved in the contraction of muscle. They are also an integral part of the unique, highly organized structure of muscle tissue.[Citation9] The same proteins can be used to give genetic evidence that cultured species are highly structured.[Citation10]
Information regarding protein polymorphism in fish germplasm is not available in our country and demands its exploration. The development of molecular markers, which are based on polymorphism observable in protein or DNA, has greatly facilitated research in phylogeny, ecology, animal breeding, and genetics. The genetic variation and structure of proteins between cultured and wild L. rohita and C. mrigala based on muscles proteins profile in different weight groups will serve as the baseline for such work in our country. Many researchers have studied genetic diversity among fish genotypes by applying DNA fingerprinting using different techniques, such as restriction length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), sequence characterized amplified region (SCAR), simple sequence repeat (SSR) markers, and randomly amplified polymorphic DNA (RAPD) markers,[Citation11,Citation12] Electrophoretic technique of sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS-PAGE) can discriminate variants by their charge or mass. It is quite rapid and an inexpensive method and is non-dependent on prior information of fish genomes.[Citation11,Citation13] This technique has been used by many workers and is still applicable.[Citation10,Citation14,Citation15]
Freshwater fish culture practices in Pakistan and India are mainly dominated by three major species, namely, L. rohita, C. mrigala, and Catla catla. Genetic studies of the distribution of protein variations were considered as a potential source of insight into biology and volution of these species.[Citation10] Hence, the present work was undertaken to extract and isolate different kinds of proteins from the meat of wild and cultured L. rohita and C. mrigala found in Pakistan and characterize them by SDS-PAGE to explore the protein-based genetic diversity in wild and cultured fish species living in two different habitats. Collection of such data is essential for any efficient fish management program or for conservation of fresh water genotypes. Such genotypes could be used as parents for random mutagenesis or incorporated for gene recombination studies before marker assisted selection/breeding can be used for fish improvement.
MATERIALS AND METHODS
Source of Fish
Seven fishes of each weight group of C. mrigala (mrigal) and L. rohita (rohu) of both cultured and wild species were procured from Fish Seed Hatchery, Faisalabad, Pakistan. The fish were immediately transported to the laboratory live and divided into three categories based on their fresh weight (w): w1, w2, and w3 weighing 500, 800, and 1100 g, respectively. The fish were washed briefly with potable water to remove slime and dirt, and drained using a sieve. They were weighed and then filleted with a stainless steel knife. The fillets were skinned and the dorsal meat was used for protein extraction.
Protein Extraction, Purification, and Estimation
One gram of minced fish meat was homogenized by grinding it in a mortar with 2 ml of glycerol and 8 ml of sodium phosphate buffer (0.05 M) as described earlier.[Citation16] The homogenate was centrifuged at 850× g for 15 min at room temperature. For purification of proteins, the supernatant was poured in the permeable egg membrane and tied tightly. It was placed in 0.1% NaCl solution for 3 h. After that, the solution from the membrane was taken with the help of a syringe and stored in caped test tubes. The purified supernatant thus obtained was used for protein estimation by following the method of Lowry et al.[Citation17] using a spectrophotometer (HITACHI Model #UV 2800, China).
Qualitative Analysis of Proteins by SDS-PAGE
SDS-PAGE (10%) was carried out as outlined by Laemmli[Citation18] using a vertical gel electrophoresis unit (SCIE-PLAS Model #TV 50, SCIE-PLAS Ltd., UK). For determination of molecular mass of each protein, two different molecular weight marker kits were purchased from Fermentas, Lahore, Pakistan (15–170 kDa), and Novagen by Merck (10–250 kDa). The gels were stained with Coomassi blue R-250, destained with 1 N NaCl, and were photographed with a digital camera. Gel doc (VILVER LOURMAT) apparatus was used for permanent recording of the results. Molecular weights of isolated proteins were determined with the help of gel doc apparatus and Photocapt (12.4) software (Photo-Capt, Vilber Lourmat, France).
Statistical Analysis
Protein data across the studied species were scored by their presence as 1 or absence of protein bands as 0 for each category. By comparing the banding patterns of proteins of each genotype, genotype specific bands were scored and faint or invisible bands were ignored. The binary data so obtained were used to determine level of polymorphisms by dividing the polymorphic bands by the total number of bands observed. Pairwise similarity and dissimilarity matrices were formed by Euclidean coefficients of similarity using Statistica computer software (Statsoft, USA). A dendrogram was constructed by using the un-weighed pair group method with arithmetic average (UPGMA). The principal component analysis using the EIGEN and PROJ modules was also applied on binary data.[Citation19,Citation20]
RESULTS
C. mrigala
Muscle proteins of weight groups w1 and w2 of both wild and cultured (each) C. mrigala were characterized by 10% SDS-PAGE (; ). In cultured fish, six protein bands of 90, 77, 68, 45, 42, 30 kDa while in wild type fish only four protein bands of 68, 43, 30, and 19 kDa, respectively, were observed. It was noted that four protein bands (68, 45, 42, and 30 kDa) were common in both cultured and wild samples. Eight and six proteins, respectively, were present in wild and cultured C. mrigala of weight group w2 (; ). It was noted that one protein band of 107 kDa was common in both cultured and wild fish meat samples of weight group w2. The cultured fish of weight group three (w3) showed 11 more proteins as compared to the wild fish samples. In the case of cultured fish, the protein bands of 210, 140–130* (*heat shock proteins), 107, 90, 68, 55, 42, 37, and 30 kDa were observed, whereas in wild fish 107, 70, 68, 55, 42, 37, 30, and 19 kDa were present. It was observed that seven proteins bands (107, 70, 68, 55, 42, 37, and 30 kDa) were common both in cultured and wild fish meat samples of weight group w3.
Table 1 Summary of muscle proteins electrophoregram of cultured and wild Cirrhinus mrigala of three weight groups
L. rohita (Rohu)
Muscle proteins of cultured and wild L. rohita of the three weight groups were also characterized by SDS-PAGE. The data of cultured and wild fish of weight group w1category are presented in . It has been observed that the cultured fish had more proteins (seven bands) as compared to wild fish (six bands) (). The proteins bands resolved from the muscles sample of cultured fish had a molecular mass of 190, 140, 90, 68, 42, 35, and 15 kDa, respectively. In the case of wild fish, the resolved proteins possessed a molecular mass of 150, 100, 77, 42, 30, and 24 kDa, respectively. It was noted that two proteins of 42 and 15–24 kDa were common in both cultured and wild fish samples in weight group w1, whereas in weight group w2, nine and four bands were resolved from cultured and wild fish, respectively, against the marker (10–250 kDa) (; ). The samples of cultured and wild fish of weight group w3 are presented in and a summary of the protein bands resolved is shown in .
Figure 2a Protein profile of cultured and wild Cirrhina mrigala of three weight groups on 10% SDS-PAGE (Lane [1–6] fish sample and marker [M]).
![Figure 2a Protein profile of cultured and wild Cirrhina mrigala of three weight groups on 10% SDS-PAGE (Lane [1–6] fish sample and marker [M]).](/cms/asset/4c718976-dc68-47a3-bb4e-3adf6adef9c4/ljfp_a_511752_o_f0002g.gif)
Figure 2b Protein profile of cultured and wild Labeo rohita of three weight groups on 10% SDS-PAGE (Lane [1–6] fish sample and marker [M]).
![Figure 2b Protein profile of cultured and wild Labeo rohita of three weight groups on 10% SDS-PAGE (Lane [1–6] fish sample and marker [M]).](/cms/asset/aedd565f-c6b4-4faa-a2a1-00b71b629bf2/ljfp_a_511752_o_f0005g.gif)
Table 2 Summary of muscle proteins electrophoregram of cultured and wild Labe rohita of three weight groups
The six cultured and wild samples in triplicate against the marker (10–170 kDa) were run on SDS-PAGE, respectively. showed that cultured fish had more proteins (11) than those of wild ones (7). In cultured and wild fish, isolated proteins were identified on the basis of their molecular masses, with the help of Biotechnology Explorer Protein Fingerprinting Instruction Manual (Catalog Number 166-0100EDU), as myosin1 and myosin2, C protein, α-actinin, meromyosin light chain, heat shock protein, γ-glubulin heavy chain, actin, myosin light chain, and troponin I, which had molecular masses of 190, 170, 146, 140, 100, 77, 62, 50, 42, 24, and 19 kDa, respectively; whereas in case of wild fish, myosin heavy chain, C protein, gelsolin, fimbrin, desmin, actin, and troponin T with molecular masses of 210, 170, 140, 115, 90, 68, 55, 42, and 30 kDa, respectively, were recognized. It was noted that three proteins (170 kDa), C protein (140 kDa), and actin (42 kDa) were common in both cultured and wild fish samples.
Cluster Analysis
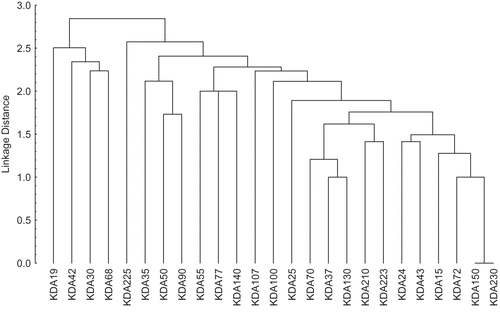
Euclidean dissimilarity coefficient matrix was calculated among wild and cultured fish samples of C. mrigala. With respect to molecular mass, their dissimilarity coefficient ranged from 1.00 (72 vs. 230 kDa, 72 vs. 150 kDa, 37 vs. 130 kDa, 15 vs. 72 kDa, and 37 vs. 70 kDa) to 3.46 (42 vs. 43 kDa, 25 vs. 30 kDa, and 24 vs. 30 KDa), respectively (). Seventeen clusters were observed on the basis of average distance (). The cluster diagram revealed the presence of 17 clusters at 50% linkage distance. Out of them, 14 clusters consisted of single proteins (19, 42, 30, 68, 225, 35, 50, 90, 55, 77, 140, 107, 100, and 25 kDa each, respectively). There were three proteins (70, 37, and 130 kDa) in cluster number 15 and two proteins (210 and 223 kDa) in cluster number 16. The 17th cluster was the largest cluster with six proteins (24, 43, 15, 72, 150, and 230 kDa). This cluster showed no difference between 150 and 230 kDa protein subunits. These results indicate that, on the basis of muscle protein, a lot of genetic variations were present in the wild and cultured fish samples. Fish weight dissimilarity coefficient ranged from 1.73 (wild fish of weight group w2 vs. cultured fish belonging to the same weight group) and in weight group w3, it was calculated as 3.8. The cluster diagram revealed the presence of eight clusters at 75% linkage distance. Out of these clusters, three clusters had a single type of fish in weight category w3. These results showed that, on the basis of fish weight, a lot of genetic variation was present in the wild and cultured fish samples.
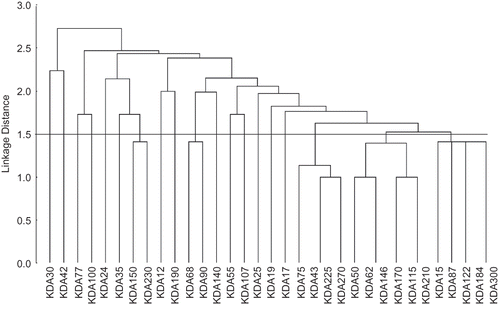
Euclidean dissimilarity coefficient matrix was calculated between cultured and wild L. rohita fish meat as well. The dissimilarity coefficient ranged from 0.00 (184 vs. 300 kDa, 43 vs. 225 kDa, 115 vs. 210 kDa, and 62 vs. 146 kDa) to 3.46 (30 vs. 35 kDa), respectively (). Nineteen clusters were observed on the basis of average distance. A phenogram for protein was constructed on the basis of UPMGA. The cluster diagram revealed the presence of 19 clusters at 50% linkage distance. Out of 19 clusters, 14 clusters contained a single protein each (30, 42, 77, 100, 24, 35, 12, 190, 140, 55, 107, 25, 19, and 17 kDa, respectively). There were two proteins (150 and 230 kDa) in cluster number 7 and two proteins (68 and 90 kDa) in cluster number 10, respectively. Cluster 17 consisted of four proteins (75, 43, 225, and 270 kDa), among them 43 and 225 kDa proteins were common. Cluster 18 was the largest cluster with six proteins (50, 62, 146, 170, 115, and 210 kDa). This cluster showed no difference between 62 and 146 kDa and between 115 and 210 kDa proteins, respectively. The 19th cluster consisted of five proteins (15, 87, 122, 184, and 300 kDa); among them 184 and 300 kDa proteins were similar. These results showed that, on the basis of proteins profile, there was quite significant genetic variation in the wild and cultured fish as weight dissimilarity coefficient ranged from 2.45 (w1) to 4.12 (w3).
The cluster diagram revealed the presence of nine clusters at 77% linkage distance. Out of nine, five clusters (I, II, IV, V, VIII) had a single type of fish weight (w3, w2, and w3). Two clusters (VI, VII) harbored two wild fish specimens only (w2 and w1) and one cluster (IX) had cultured fish specimens (w2) only. Cluster 3 was the largest with three specimens, two cultured and one wild type consisting of fish of different weights (w2) (). Association among all genotypes was also resolved by using principle component analysis (PCA) (results not shown). The overall grouping pattern of PCA was quite similar to that shown in the dendrogram.
DISCUSSION
Farm animal genetic diversity is required to meet current production needs in various environments to allow sustained genetic improvement and to facilitate rapid adaptation to changing breeding objectives. The current study was designed in order to get a better understanding of genetic diversity among cultured and wild C. mrigala and L. rohita in Pakistan.
This aspect has received little attention in our country. Muscle protein electrophoregram revealed by SDS-PAGE was used as a tool for phylogenetic analysis. Electrophoretic muscle protein profiles for all fish samples were noted on the basis of absence (0) and presence (1) of protein bands in the fish samples. On the basis of band pattern information, data was tabulated. Data showed that the protein profile in the case of wild fish samples revealed overall 25 proteins ranging from 15 to 225 kDa (). According to molecular weights, 30 kDa proteins were present in most of the fish samples (five out of seven), whereas unique protein bands (140, 90, 77, 72, 70, 43, 37, 24, and 15 kDa) were observed in different weight group samples. In wild and cultured C. mrigala of weight group w1, four and six proteins were recorded. The maximum proteins (8/25) were observed in the case of wild fish samples of weight group w3. The protein profile in the case of cultured fish samples revealed overall 26 proteins ranging from 15 to 230 kDa. According to the molecular weight, 42 kDa protein bands were present in all cultured fish samples (seven out of seven), whereas a unique protein band of 230, 223, 150, 130, 100, 72, 37, and 33 kDa was observed in different weight samples. In the case of wild C. mrigala of weight group w3, the maximum number of proteins (11/26) was present.
In wild and cultured L. rohita of weight group w1, a minimum number (4 and 6) of proteins was present. The maximum number of proteins (9/30) was observed in the case of a wild fish sample of weight group w3. showed the protein profile in the case of cultured fish samples, which revealed overall 31 proteins ranging from 10 to 300 kDa. According to molecular mass, 35, 42, and 77 kDa proteins were observed in most of the fish specimens (five out of seven), whereas the minimum number of protein (one) was observed in the case of 300, 225, 184, 170, 146, 75, 62, 55, 30, and 15 kDa, respectively, in different weight fish samples. In cultured fish samples of weight group w2, eight proteins were observed, whereas the maximum number of proteins (11/31) were observed in the case of cultured fish sample of maximum weight group, i.e., w3.
In terms of qualitative analysis it was observed that, irrespective of the same concentration of protein, there was a significant difference in protein electrophoresis among different groups of cultured and wild fish ( and ; and ). Our findings are in line with the finding of Jasra et al.[Citation21] for myofibril proteins. They isolated myosin light chain, tropomyosin, Troponin-C and m-protein specifically from the muscle fibers of two freshwater fish species, viz., Heteropneustes fossils (Bloch) and L. rohita (Hamilon). Kim et al.[Citation22] reported that the major sarcoplasmic bands were 43, 40, 17, and 11 kDa in rockfish (Sebastes fiavidus). In the present case, a 43-kDa band is present in the wild sample of C. mrigala. Okagaki et al.[Citation23] reported that molecular weight of light chains of myosin of carp ranged between 16 and 26 kDa. The presence of light chain myosin, troponin, and parvalbumin in muscle of L. rohita are in line with the findings of Montowska and Pospiech[Citation24] and Lech et al.[Citation25] Fock and Hinssen[Citation26] examined C-protein in carp skeletal muscle, while in the present study, C-protein is also found in the muscle of both cultured and wild L. rohita (data not shown). Huriaux[Citation27] detected two myosin LC2 chains and two troponin (I) isoforms through polyacrylamide gel electrophoresis from Brycon moorie. Nebulin was isolated and characterized in L. rohita as an integral part of skeletal muscle of fish as reported earlier.[Citation26,Citation27]
In the case of C. mrigala, samples revealed the presence of a lot of protein variation in both categories. On the basis of fish weight, three clusters were generated from each fish (wild or cultured), which suggested the uniqueness of these clusters. This might be due to the presence of characteristic types of proteins or because of approximately equal weight isoforms. A wide range of Euclidean distance in fish weight and protein is also one of the indications for the presence of genetic variation among these fish samples. While comparing cultured fish with the wild, it is found that in cultured fish 42 kDa protein subunit is present in all samples, whereas in wild samples 30 kDa protein subunit is the most abundant (five out of seven). However, in cultured fish in contrast with wild fish, 77 and 35 kDa proteins were present in abundance. The number of proteins increased from lower to higher weight category of fish in both cases (cultured and wild), which supported our assumption that the qualitative variation may exist in these freshwater fish species with an increase in body weight. The overall comparison between the two habitats of C. mrigala and L. rohita indicated that more numbers of proteins were present in the cultured fish as they were reared on artificial feed that was rich in proteins while wild fish were devoid of essential nutrients.
CONCLUSION
The present study has revealed the genetic heterogeneity among cultured and wild L. rohita and C. mrigala. It was noted that wild fish showed more genetic variation as compared to cultured fish that could be due to unmonitored breeding and intervention of other fish species in the wild habitat. Deterioration of anthropogenic habitat and its fragmentation, leads not only to extinction of fish species but also causes devastating influences on genetic and evolutionary consequences. To meet the growing demand of fish consumers, policy makers should advise the fish farmers that they should manage connectivity between fragmented habitat and spatially distinct populations in wild habitat so that genetic variability in the cultured species can be conserved. This study has shown for the first time in Pakistan that protein-based genetic variation increases with weight group, which could be due to the maturity of fish to support formation of age expression-related proteins that provide material for genetic analysis. This analysis has allowed temporal genetic variation in these populations. Recently, protein electrophoresis-based genetic evaluation has been superseded by DNA-based studies,[Citation10,Citation11] but the authors' data sets may continue to provide guidelines for testing genetic changes along with DNA marker-assisted genome studies.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the Higher Education Commission of Pakistan and British Council, Pakistan for providing funding under JHELP. The authors also wish to acknowledge the School of AFRD, University of Newcastle, UK for providing technical training to the Link Staff under the JHELP program.
REFERENCES
- Dey , M.M. , Rab , M.A. , Paraguas , F.J. , Piumsombun , S. , Bhatta , R. , Alam , M.F. and Ahmed , M. 2005 . Fish consumption and food security: A disaggregated analysis by types of fish and classes of consumers in selected Asian countries . Aquaculture Ecology and Management , 9 : 89 – 111 .
- Mackie , I.M. 1994 . “ Fish protein ” . In New and Developing Sources of Food Proteins , Edited by: Hudson , B.J.F. 95 – 143 . New York : Chapman and Hall .
- Sathivel , S. , Huang , J. and Bechtel , P.J. 2008 . Properties of Polloc (Theragra chalcogramma) skin hydrolysates and effects on lipid oxidation of skinless Pink Salmon (Oncorhynchus gorbuscha) fillets during four months of frozen storage . Journal of Food Biochemistry , 32 : 247 – 263 .
- Zibaee-Nezhad , M.J. , Khosravi , M. , Akbari , S.N. , Bani-Asadi , N. and Affilia , E.G. 2010 . Omega-3 fatty acid composition of Persian Gulf fishes . International Journal of Food Properties , 13 : 574 – 579 .
- Memon , N. , Talpur , F.N. and Bhangar , M.I. 2010 . A comparison of proximate composition and fatty acid profile of Ibndus river fish species . International Journal of Food Properties , 13 : 328 – 337 .
- Sahin , S. and Sumnu , G. 2001 . Effect of microwave cooking on fish quality . International Journal of Food Properties , 4 : 501 – 512 .
- Xiong , Y.L. 1997 . “ Structure-function relationship of muscle proteins ” . In Food Proteins and Their Applications , Edited by: Damodaran , S. and Paraf , A. 341 – 392 . New York : Marcel Dekker .
- Kolanowski , W. 2010 . Omega-3 LC PUFA contents and oxidative stability of encapsulated fish oil dietary supplements . International Journal of Food Properties , 13 : 498 – 511 .
- Skaara , T. and Regenstein , J.M. 1990 . The structure and properties of myofibrillar proteins in beef, poultry and fish . Journal of Muscle Foods , 1 : 269 – 291 .
- Verspoor , E. , Beardmore , J.A. , Consuegra , S. , DeLeaniz , C.G. , Hindar , K. , Jordan , W.C. , Koljonen , M.-L. , Mahkrov , A.A. , Paaver , T. , Sánchez , J.A. , Skaala, Titov , S. and Cross , T.F. 2005 . Population structure in the Atlantic salmon: Insights from 40 years of research into genetic protein variation . Journal of Fish Biology , 67 : 3 – 54 .
- Anne , C. 2006 . Choosing the right molecular genetic markers for studying biodiversity: from molecular evolution to practical aspects . Genetica , 127 : 101 – 120 .
- Borroso , R.M. , Hilsdorf , A.W.S. , Moreira , H.I.M. , Cabello , P.H.C. and Traub-Cseko , Y.M. 2005 . Genetic diversity of wild and cultured population of Brycon opalinus (Cuvier, 1819) (Characiforme, characidae, Bryconiae) using microsatellites . Aquaculture , 247 : 51 – 65 .
- Zawandzki , C.H. , Ronesto , E. , de Paiva , S. and Lara-Kamei , C.S. 2004 . Allozyme differentiation of four populations of Hypostomus (Teleostei:oricariidae) from Ribeirao Keller, a small stream in the upper Rio Parana basin, Brazil . Genetica , 121 : 251 – 257 .
- Mitsuhashi , T. , Kasai , M. and Hatae , K. 2002 . Detection of giant myofibrillar proteins connectin and nebulin in fish meat by electrophoresis in 3–5 gradient sodium dodecyle sulfate polyacrylaminde slab gel . Journal of Agricultural and Food Chemistry , 50 : 7499 – 7503 .
- Okagaki , T. , Takami , M. , Josokawa , K. , Yano , M. , Fujime , S. and Hooi , A. 2005 . Biochemical properties of ordinary and dark muscle myosine from carp skeletal muscle . Journal of Biochemistry , 38 : 255 – 262 .
- Hultin , H.O. and Kelleher , S.D. Process for isolating a protein composition from a muscle source and protein composition . U.S. Patent 6,005,073 . December 21, 1999 .
- Lowry , O.H. , Roserough , N.J. , Farr , A.L. and Randall , R.J. 1951 . Protein measurement with the Folin phenol reagent . Journal of Biological Chemistry , 193 : 265 – 275 .
- Laemmli , U.K. 1970 . Cleavage of structural proteins during the assembly of the head of bacteriophage T4 . Nature , 227 : 680 – 685 .
- Rohlf , F.J. 2002 . NTSYS.pc: Numerical taxonomy system ver. 21 , Setauket , New York : Exeter Publishing Ltd .
- Ward , R.D. , Skibinski , D.O.F. and Woodward , M. 1992 . Protein heterogeneity, protein structure and taxonomic differentiation . Evolutionary Biology , 26 : 73 – 159 .
- Jasra , P.K. , Talesara , C.L. and Kiran , S. 1991 . Polymorphism of myofibrillar proteins in histochemically identified myotomal muscle fibre types of Heteropneusts fossilis (Bloch) and Labeo rohita (Hamilton) . Journal of Fish Biology , 38 : 165 – 173 .
- Kim , Y.S. , Yougsawatdigul , J. , Park , J.W. and Thawornchinsombt , S. 2005 . Characteristics of sarcoplasmic proteins and their interaction with myofibrillar proteins . Journal of Food Biochemistry , 29 : 517 – 532 .
- Okagaki , T. , Suzuki , R. and Ooi , A. 2007 . C-protein (MyBP-C) isoforms from carp ordinary and dark muscles and muscle type-specific binding to myosin . Fisheries Science , 73 : 640 – 650 .
- Montowska , M. and Pospiech , E. 2007 . Species identification of meat by electrophoretic method . ACTA methods. ACTA Scientiarum Polonorum Technologia Alimentaria , 6 : 5 – 16 .
- Lech , J.J. , Lewis , S.K. , Friedman , M.A. , Johnson , L.A. and Mend Mueller , L.M. 1996 . Binding of acrylonitrile to parvalbumin . Toxicological Science , 29 : 260 – 266 .
- Fock , U. and Hinssen , H. 2002 . Nebulin is a thin filament protein of the cardiac muscle of the agnathans . Journal of Muscle Research and Cell Motility , 23 : 205 – 213 .
- Huriaux , F. , Baras , E.S. , Vandewalle , P. and Focant , B. 2003 . Expression of myofibrillar proteins and parvalbumin isoforms in white muscle of Dorada during development . Journal of Fish Biology , 62 : 774 – 792 .