Abstract
The objective of the present study was to assess the potential of front-face fluorescence spectroscopy coupled with chemometric tools for the evaluation of the quality of milk samples according to the feeding system and genotype. Fifty (n = 50) ewe's milk samples were scanned after excitation set at 250, 290, 322, and 380 nm and emission set at 410 nm. Thirty out of the 50 samples composed the first trial and were obtained from two different genotypes (i.e., Comisana versus Sicilo-Sarde); the second trial was composed of 20 samples obtained from the Sicilo-Sarde genotype with two different feeding systems in pen (soybean versus scotch bean). Milk samples were divided into four groups named Sicilo-Sarde with pasture feeding (Spas), Comisana with pasture feeding (Cpas), Sicilo-Sarde feeding on scotch bean (Ssco), and Sicilo-Sarde feeding on soybean (Ssoy). The factorial discriminant analysis was applied to the: (i) four groups (i.e., Spas, Ssco, Ssoy, and Cpas) and (ii) three groups composed only of Sicilo-Sarde genotype (i.e., Spas, Ssco, and Ssoy). Considering the four groups, the best result was obtained with the excitation vitamin A spectra since correct classification amounting to 76% was observed. When the factorial discriminant analysis was performed with the three groups belonging to the Sicilo-Sarde genotype, the best result was obtained again with vitamin A spectra (i.e., emission and excitation spectra) since 88.6% of correct classification was observed. Concatenation technique applied to the five fluorescence spectra improved the rate of classification between the four groups since 44 out of 50 samples were correctly classified. No misclassification was observed between milk samples collected from ewes with pasture feeding from the pen feeding. It was concluded from the obtained results that fluorescence spectroscopy could be considered as a powerful tool for differentiating between raw milks according to both genotype and feeding system.
INTRODUCTION
The dairy ovine breeding in Tunisia is made up mainly of the Sicilo–Sarde herd, which is found, almost exclusively, in the north of the country, particularly, in the areas of Béja and Bizerte; where the climatic conditions allow the production of both fodder and pasture. The Sicilo–Sarde livestock is the result of crossbreeding between Sarde and Comisana, whose origin is probably Sicily. Comisana sheep have been introduced for the first time in Tunisia in 1991. The two breeding ewes are considered among the best dairy ovine races in the Mediterranean basin.[Citation1]
Although intensification of livestock in Tunisia is increasing, its integration with crops is limited. Integration of forage into rain-fed farming has not really succeeded. The cultivated forage area has remained constant during the last decade and its contribution to animal feeding is limited.[Citation2] However, livestock amounts have significantly increased, especially, dairy ewes. Moreover, the contribution of natural pasture and range in the feed calendar was dramatically decreased due to frequent drought and overgrazing. To better cope with this problem and to meet animal requirements, increasing amounts of feedstuffs, namely soybean meal, are imported (270.3 and 391.2 tonnes in 2003 and 2004, respectively) from some western countries, which weigh down the national economy ($61.9 million in 2003 to $126.3 millions in 2004).[Citation3]
In the last few years, the economic world conjuncture involved rising prices of soybean meals constituting the basic raw materials in concentrated food formulations for Tunisian livestock. Therefore, research for better alternatives that involves partial or total replacement of soybean meals with local food resources is essential. Although soybean meal has been well established as the main protein source for animal nutrition[Citation3] scotch bean is recently becoming available in Tunisia as a local and cost effective product. Consequently, it could be used as a substitute for soybean meal. As a protein supplement in rations of growing and fattening sheep, previous research studies have shown that scotch bean meals gave similar gain and feed efficiency as soybean meal did.[Citation4]
Recently, some research studies have assessed the potential of visible near infrared (Vis-NIR) spectroscopy to predict some chemical parameters of ewe's milk with pasture, soybean, and scotch bean feeding of two genotypes (Comisana and Sicilo-Sarde). Among the investigated parameters, good prediction of protein, ash, lactose, and freezing point were observed, while unsuccessful results (i.e., only discrimination between high and low values) were observed for pH and fat values suggesting the non usefulness of Vis-NIR for the prediction of these two parameters for ewe's milk.[Citation5]
Front face fluorescence spectroscopy (FFFS) has been used for: (i) monitoring milk samples during coagulation;[Citation6, Citation7] (ii) differentiating between cows milk samples originating from different origins;[Citation8] (iii) discriminating between milk samples according to the feeding system[Citation9 − Citation11] and genotype;[Citation12] identifying different heat treatments (pasteurisation, high pasteurisation, direct UHT, indirect UHT, and sterilisation) of 200 commercial milk samples stored at 25 and 35°C for 90 days.[Citation13]
Although, some research studies were recently performed to determine the quality of ewe's raw milk following the use of Vis-NIR, there is no study, to the best of our knowledge, on using FFFS for differentiating simultaneously milk samples according to the feeding systems and genotypes. Thus, the objective of this present study was to determine the usefulness of FFFS to discriminate between 50 ewe's milk samples according to the feeding system (pasture, soybean, and scotch bean meal) and genotype (Sicilo-Sarde versus Comisana).
MATERIALS AND METHODS
The study was divided into two distinct experiments. The objectives of the first and second experiments were to determine, respectively, the effect of the: (i) feeding system and (ii) genotype on the quality of the milk.
Experiment 1
Twelve 5-year-old Sicilo-Sarde ewes weighing 42.2 kg on average, at their third lambing were kept at 17°C in individual boxes and in an environmentally controlled sheepfold. Zootechnical performances and energy intake were recorded individually. After 2 weeks of adaptation, ewes were divided into two homogenous weight matched groups (n = 6). The two experimental diets differed in terms of protein sources (soybean versus scotch bean) as illustrated in and . Ewes remained on the experiment until 10 weeks post-partum. Diets were iso-energetic and were given in restricted amounts according to the feed intake of the two groups.
Experiment 2
Two genotypes, Comisana ewes (n = 156, weight = 45 kg) aged 4.3 years old and Sicilo-Sarde ewes (n = 50, weight = 50 kg) aged 5.2 years old were used. Animals were maintained on pasture feeding (yearly pasture of barley, oat, and clover or permanent pasture of ray grass) and supplemented with oat hay and concentrate (34 corn, 17 wheat, 19 soya, 25 barley, and 5 g 100 g−1 mineral and vitamin premix). They were inspected by a qualified shepherd on a daily basis, and routine animal care and vaccination procedures were conducted as prescribed by best practice protocols. Ewes remained on the experiment until 15 weeks post-partum.
All the analyses were determined at 7-day intervals during 10 (experiment 1) and 15 (experiment 2) weeks lactation period. For each group, the milk samples collected from the different ewes (n = 6 per group for experiment 1; and n = 156 and 50 for experiment 2) were mixed and an aliquot of 100 ml was taken and kept in a freezer at −40°C until analyses. Before each analysis, milk samples were thawed during the night at 4°C in a refrigerator. Thus, the number of the analysed samples was 50 samples (10 samples for each group of experiment 1 and 15 samples for each group of experiment 2).
Physico-Chemical Parameter and Fluorescence Measurements
Milk samples were analysed for pH, density, dry matter, fat, protein, lactose, ash, and freezing point by using LactoScan (Milkotronic LTD, Serial no. 4696, Stara Zagora, Hungary). Fluorescence spectra were recorded using a Fluorolog-2 spectrofluorimeter (Spex-Jobin Yvon, Horiba, the Netherlands) mounted with a variable angle front-surface accessory. The incidence angle of the excitation radiation was set at 22.5° to ensure that reflected light, scattered radiation, and depolarisation phenomena were minimised. The emission spectra of aromatic amino acids and nucleic acids (AAA+NA) (280–450 nm; increment 1 nm), tryptophan residues (305–450 nm; increment 1 nm), vitamin A (330–540 nm; increment 1 nm), and riboflavin (400–640 nm; increment 1 nm) were recorded with the excitation wavelengths set at 250, 290, 322, and 380 nm, respectively. The excitation spectra of the vitamin A (280–350 nm; increment 1 nm) were acquired with the emission wavelength set at 410 nm. All spectra were corrected for instrumental distortions in excitation using a rhodamine cell as a reference channel.
Table 1 Diet formulation (a) and chemical composition (b) of the rations given to the ewes feeding in pen during 10 weeks of lactation period.(a)
Statistical Analyses
In order to reduce scattering effects and to compare milk samples, fluorescence spectra were normalised by reducing the area under each spectrum to a value of 1 according to Bertrand and Scotter.[Citation14] Mainly the shift of the peak maximum and the peak width changes in the spectra were considered following this normalisation. The principal component analysis (PCA) was applied to the normalised spectra to investigate differences among milk samples. The PCA transforms the original variables into new axes called principal components (PCs), which are orthogonal, so that the data set presented on these axes are uncorrelated with each other. Therefore, PCA explains the total variation found in the data set in only a few PCs and each successively derived PC expresses decreasing amounts of the variance. This statistical multivariate treatment was earlier used to observe similarities among different milk samples,[Citation8] reducing the dimension to two or three PCs, while keeping most of the original information found in the data sets.
In a second step, FDA was performed separately on the first 10 PCs scores of the PCA of each excitation and emission spectra, which contains the whole information found in raw data sets. The aim of this technique is to predict the membership of an individual to a qualitative group defined previously.[Citation15] A group was created for each type of milk, i.e., milk sample from Sicilo-Sarde ewes feeding scotch bean (Ssco), milk samples from Sicilo-Sarde ewes feeding soybean (Ssoy), milk samples from Sicilo-Sarde ewes with pasture feeding (Spas), and milk samples from Comisana ewes with pasture feeding (Cpas). The method cannot be applied in a straightforward way to continuous spectra because of the high correlations occurring between the wavelengths. Advantages were found in the preliminary transformation of the data into their PCs scores. FDA assesses new synthetic variables called “discriminant factors,” which are linear combinations of the selected PCs, and allows better separation of the centres of gravity of the considered groups. The individual milk samples can be reallocated within one of the four groups (Ssco, Ssoy, Spas, or Cpas) or the three groups when only the Sicilo-Sarde ewe's milk were considered (Ssco, Ssoy, or Spas). For each milk sample, the distance from the various centres of gravity of the groups is calculated. The milk sample is assigned to the group where its distance between the centre of gravity is the shortest. Comparison of the assigned group to the real group is an indicator of the quality of the discrimination.
With leave one-out cross-validation, the same sample is used both for calibration and validation model. A sample is left out from the calibration data set and the model is calibrated on the remaining data points. Then the value for the left-out sample is predicted and the prediction residual is computed. The process is repeated with another sample of the calibration set, and so on until every sample has been left out once; then all prediction residuals are combined to compute the validation step.
Finally, the first ten PCs scores of the PCA performed on each of the fluorescence spectral data set were pooled into one matrix (concatenation) and this new table was analysed by FDA.[Citation16] The process consists of putting one beside the other in the same matrix using the first ten PCs of the PCA resulting from each excitation and emission fluorescence spectra. The aim of the concatenation technique was to take into account the information contained in the fluorescence spectra recorded at different excitation and emission spectra allowing improvement of the discrimination of the investigated milk samples according to the feeding system and genotype. PCA was performed under DOS by using the “Saisir” package of D. Bertrand (INRA, Nantes) and the FDA was performed using StatBoxPro (Grimmer Logiciels, Paris, France).
RESULTS AND DISCUSSION
Physico-Chemical Analyses
From , significant difference between the feeding systems (pasture feeding versus pen feeding) was observed regardless of the genotype (Comisana and Sicilo-Sarde). Considering milk samples belonging to Cpas and Spas groups, no significant difference between all the investigated physico-chemical parameters was observed indicating that ewe's genotype did not present a significant effect on the physico-chemical parameters (P ≥ 0.05). Similar results were obtained for milk samples originating from Ssco and Ssoy groups, excepting fat amount where the highest values were observed for milk samples from Ssoy group (7.85 versus 6.75 g 100 g−1).
Table 2 Average values of the physico-chemical composition of milk samples from Sicilo-Sarde ewes feeding on soybean meal (Ssoy), Sicilo-Sarde ewes feeding on scotch bean (Ssco), Sicilo-Sarde ewes with pasture feeding (Spas), and Comisana ewes with pasture feeding (Cpas) during the lactation period
Fluorescence Measurements
The emission spectra of AAA+NA and tryptophan fluorescence spectra showed similar trends (data not shown). A maximum located around 347–348 nm was observed for all feeding systems, genotypes, and lactation stages considered in this study. Quite similar results were obtained on the emission spectra of vitamin A (data not shown), which presented a maximum around 408–409 nm.
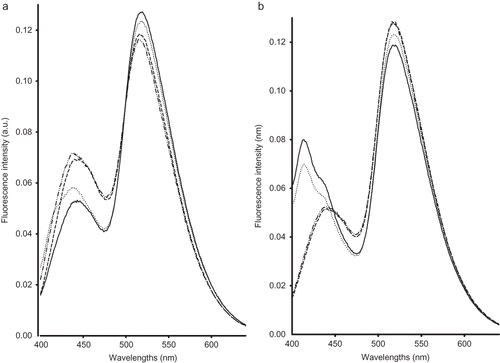
Regarding riboflavin spectra, a clear discrimination between milk samples according to the feeding system and genotype was observed, regardless of the lactation stage (and ). All the investigated milk samples showed two maxima located around 440 and 520 nm, except milk samples of Cpas and Spas groups at the end of the lactation period (i.e., 10 for experiment 1 and 15 weeks for experiment 2), which presented two bands around 520 and 416 nm and a shoulder at 440 nm (). The peak at 520 nm has been attributed to riboflavin,[Citation17] while the peaks located around 416 and 440 nm have not yet been identified. seems to demonstrate that milk samples collected from Spas and Cpas groups were less oxidised than those belonging to the two other groups in agreement with the findings of Hammami et al.;[Citation9] however, at the end of the lactation stage, an opposite trend was observed.
Figure 1 Normalised emission riboflavin fluorescence spectra recorded following excitation at 380 nm on milk samples at the beginning (a) and end (b) of lactation stage of Comisana with pasture feeding (—), Sicilo-Sarde with pasture feeding ( … ), Sicilo-Sarde feeding on scotch bean (− − −), and Sicilo-Sarde feeding on soybean (— .. — ..).
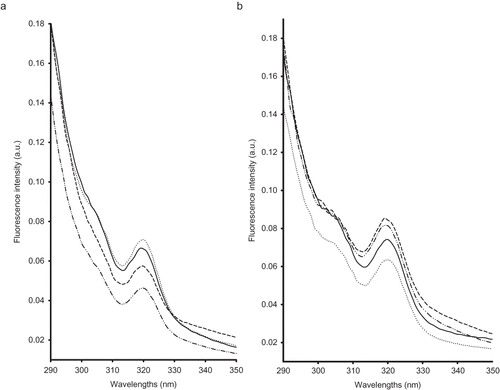
The vitamin A excitation spectra showed a maximum and a shoulder around 320 and 305 nm, respectively. From , milk samples originating from Spas and Cpas groups differed from those being fed in pen (Ssco and Ssoy groups). This difference could be due to the interferences of other milk components with vitamin A that could affect the shape of the spectra (i.e., protein-lipid interactions);[Citation8, Citation9, Citation11, Citation12] indeed, at week 1, milk samples belonging to Spas and Cpas groups presented the highest level of fat (i.e., ˜11 g 100 g−1) while milk samples originating from the two other groups presented the lowest one (i.e., 7.1 and 7.93 g 100 g−1 for Ssco and Ssoy groups, respectively).
Figure 2 Normalised excitation vitamin A fluorescence spectra recorded following excitation at 410 nm on milk samples at the beginning (a) and end (b) of the lactation stage of Comisana with pasture feeding (—), Sicilo-Sarde with pasture feeding ( … ), Sicilo-Sarde feeding on scotch bean (– – –), and Sicilo-Sarde feeding on soybean (— .. — ..).
From and , vitamin A spectra showed the largest difference between milk samples compared to the other fluorescence spectra. However, due to the complexity of the investigated spectra, univariate analysis was not appropriate to statistically analyse the data sets. Multivariate statistical techniques, such as PCA and FDA, made it possible to extract information related to the environment of the intrinsic probes.
Milk discrimination based on aromatic amino acids and nucleic acids and tryptophan fluorescence spectra throughout the lactation stage
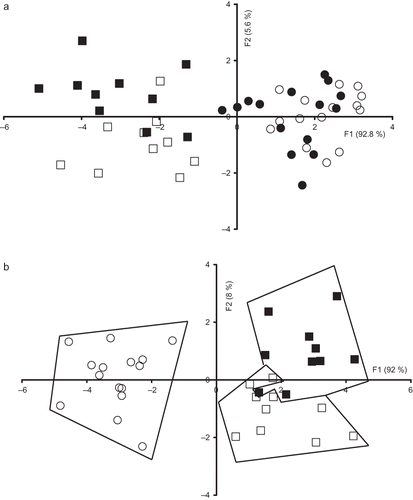
FDA was applied on the first ten PCs scores of the PCA performed on each spectral data set. Before applying the FDA, four milk groups were created (Ssco, Ssoy, Spas, and Cpas). The map defined by the first two discriminant factors taking into account 98.4% of the total variation was shown . The discriminant factor 1 accounting for 92.8% of the total variation allowed a good discrimination between milk samples belonging to Spas and Cpas from the two other groups. Although a trend to a good discrimination between milk samples originating from Ssco and Ssoy groups was observed according to the discriminant factor 2, Cpas and Spas groups were overlapped on the map. From the obtained result, it could be concluded that AAA+NA spectra are more sensitive to the feeding system (pen versus pasture) than to the genotype. This assumption was confirmed by the classification rate illustrated in since only 50% of samples were correctly classified. From this table, it was observed that no milk sample belonging to Ssco and Ssoy groups was misclassified with Spas and Cpas groups. The table shows also that milk samples belonging to ewes fed on pasture (Spas and Cpas) were not misclassified with milk samples belonging to ewes fed in pen (Ssco and Ssoy), excepting one sample belonging to Cpas group, which was found to be assigned to Ssoy group.
Figure 3 Discriminant analysis similarity map determined by discriminant factors 1 (F1) and 2 (F2) for the factorial discriminant analysis (FDA) performed on the emission aromatic amino acids and nucleic acids spectra with leave one-out cross-validation of: (a) Comisana with pasture feeding (•), Sicilo-Sarde with pasture feeding (º), Sicilo-Sarde feeding on scotch bean (▪), and Sicilo-Sarde feeding on soybean (□) groups; and (b) Sicilo-Sarde with pasture feeding (º), Sicilo-Sarde feeding on scotch bean (▪), and Sicilo-Sarde feeding on soybean (□) groups.
Regarding tryptophan fluorescence spectra, the first two-discriminant factors and any other discriminant factors did not allow a clear differentiation between the four groups. Most of the samples were overlapped on the map (data not shown). A total of 52% of samples were correctly classified (). From this table, most of milk samples belonging to one group were misclassified with the three other groups. It can be concluded that tryptophan fluorescence spectra cannot be used for the differentiation between milk samples according to the genotype and/or the feeding system.
Table 3 Classification table of milk samples from Sicilo-Sarde ewes feeding on soybean meal (Ssoy), Sicilo-Sarde ewes feeding on scotch bean (Ssco), Sicilo-Sarde ewes with pasture feeding (Spas), and Comisana ewes with pasture feeding (Cpas) throughout the lactation period based on aromatic amino acids and nucleic acids (AAA+NA), tryptophan, riboflavin, and vitamin A spectra and the concatenated fluorescence spectra for: (a) the four (Spas, Cpas, Ssco, and Ssoy) and (b) the three (Spas, Ssco, and Ssoy) groups.(a)
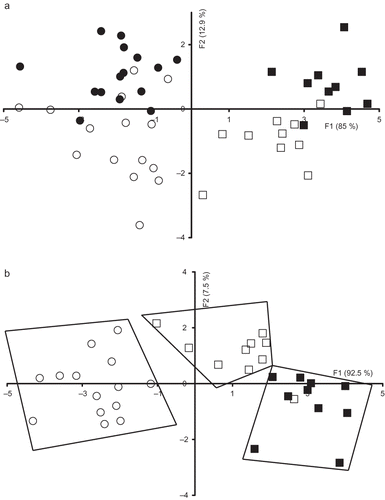
By using the same approach (i.e., FDA), the potential of AAA+NA and tryptophan fluorescence spectra to discriminate milk samples collected from ewes presenting the same genotype (Sicilo-Sarde) according to the feeding system (Ssco, Spas, and Ssoy) was determined. The map defined by the first two-discriminant factors of the FDA performed on the AAA+NA allowed a clear discrimination between the three groups (). Indeed, considering the discriminant factor 1 accounting for 92% of the total variation, Spas milk group was located on the left, while the other two groups were observed on the right. The discriminant factor 2 differentiated between Ssco groups, which had positive scores from Ssoy group exhibiting negative values.
By applying FDA to the three groups (Spas, Ssoy, and Ssco) with leave one-out cross-validation, correct classification amounting to 80 and 60% were observed, respectively, with AAA+NA and tryptophan spectra (). Regarding AAA+NA spectra, 14 out of 15 Spas milk samples were correctly classified; only 1 sample was classified as belonging to the Ssoy group, which could be explained by the supplementation of Spas group with soybean meal at a level of 19%. Seventy percent correct classification was observed for Ssco and Ssoy groups (). The relatively low percent of correct classification could be due to the relative low number of the investigated milk for the two latter groups (n = 10). Regarding tryptophan fluorescence spectra, two samples belonging to Spas group were classified as belonging to Ssoy group. For Ssco and Ssoy groups, correct classification rates of 30 and 50%, respectively, were observed indicating that tryptophan fluorescence spectra could not be used as a potential tool for differentiating between milk samples according to the feeding system ().
Milk discrimination based on riboflavin fluorescence spectra throughout the lactation stage
The FDA was applied to the first ten PCs scores of the PCA performed on fluorescence spectra obtained after excitation at 360 nm and the similarity map of the first two-discriminant factors allowed separation of milk samples belonging to Cpas and Spas groups from Ssco and Ssoy groups (data not shown). In addition, the four groups were mostly well discriminated indicating the ability of riboflavin spectra to discriminate between the investigated milk samples. Compared to the AAA+NA and tryptophan fluorescence spectra, better result was obtained with riboflavin spectra since correct classification amounting to 68% was observed (). The best classification was observed with milk samples belonging to Ssco and Ssoy groups; indeed, 80 and 90% of Ssco and Ssoy samples were correctly classified. Regarding milk samples belonging to Cpas and Spas groups, 5 and 8 samples out of 15 were respectively misclassified.
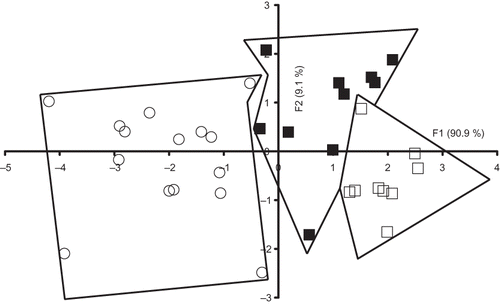
When Sicilo-Sarde ewe's milk samples were analysed alone by FDA, a good separation between the investigated groups was observed (). Milk samples belonging to Spas group presented negative scores according to the discriminant factor 1 accounting for 90.9% of the total variation, while Ssco and Ssoy groups exhibited mostly positive scores. These two latter groups were clearly separated. Correct classification amounting to 85.7% was observed for all the samples (). Two samples belonging to Ssco group and two others assigned to Spas group were misclassified giving correct classification rates of 80 and 86.7%, respectively. The best result was obtained with Ssoy group since nine out of ten samples were correctly classified. The obtained results confirmed the ability of riboflavin spectra for identifying milk samples originating from Sicilo-Sarde ewes according to the feeding system.
Figure 4 Discriminant analysis similarity map determined by discriminant factors 1 (F1) and 2 (F2) for the factorial discriminant analysis (FDA) performed on the emission riboflavin spectra with leave one-out cross-validation of Sicilo-Sarde with pasture feeding (º), Sicilo-Sarde feeding on scotch bean (▪), and Sicilo-Sarde feeding on soybean (□) groups.
Milk discrimination based on vitamin A excitation and emission fluorescence spectra throughout the lactation stage
Emission vitamin A spectra
Although, the FDA applied to the vitamin A emission spectra did not allow good discrimination between the four groups, a clear differentiation between Spas and Cpas groups from Ssco and Ssoy groups was observed according to the discriminant factor 1 accounting for 88.3% of the total variation (data not shown). Some overlapping between Spas and Cpas groups on one hand and between Ssco and Ssoy groups on the other hand was observed. This result was confirmed in . Indeed, a correct classification level of 64% was observed for all the samples. Seven milk samples belonging to Cpas group were classified as originating from Spas group; similar results were obtained for Spas group since 6 out of 15 samples were assigned to Cpas group. The most interesting result was that no milk sample belonging to Spas and Cpas groups was classified as belonging to samples belonging to Ssco and Ssoy groups. This conclusion was supported following the application of the FDA to the Sicilo-Sarde ewe's milk feeding with three different systems. Correct classification amounting to 88.6% was observed for all the 35 samples (). One sample belonging to Ssco group and another sample belonging to Spas group was classified with Ssoy and Ssco groups, respectively. Finally, two Ssoy samples were found to be assigned to Ssco group.
Excitation vitamin A spectra
The FDA performed to vitamin A excitation spectra acquired on milk samples belonging to the four groups is shown in . According to discriminant factor 1 accounting for 85% of the total variation, milk samples belonging to Spas and Cpas groups were separated from Ssco and Soy groups. The best classification was obtained with vitamin A excitation spectra since a correct classification rate of 76% was observed (). Indeed, Cpas, Ssco, Spas, and Ssoy were predicted with correct classification rates amounting to 80, 90, 66.7, and 70%, respectively. This result was confirmed following the application of the FDA to the Spas, Ssco, and Ssoy groups (). The three groups were well discriminated. Overall correct classification amounting to 88.6% was observed when the three groups belonging to Sicilo-Sarde genotype were analysed (). The Spas group was 100% correctly classified. Only one out of ten Ssco samples was assigned to Ssoy group. The worst classification was obtained with Ssoy group since three milk samples were misclassified: one sample with Ssco group and two samples to Spas group. The misclassification of Ssoy with Spas could be due to the presence of soybean meal at a level of 19% for Spas group. The obtained result suggested the ability of the excitation vitamin A spectra to be a useful probe for the evaluation of milk samples according to the feeding system and genotype, in agreement with a previous investigation reporting that excitation of vitamin A spectra could be used for the differentiation between: (i) milk samples submitted to different treatments (i.e., homogenised, heat-treated, and homogenised and heat-treated milk samples);[Citation18] (ii) Emmental cheeses originating from different countries[Citation19, Citation20] and soft cheeses at the external and central zone;[Citation21] (iii) fresh and aged egg yolk samples stored at room (˜20°C) and 12°C with atmosphere containing or not 2% of CO2.[Citation22 − Citation24]
Figure 5 Discriminant analysis similarity map determined by discriminant factors 1 (F1) and 2 (F2) for the factorial discriminant analysis (FDA) performed on the excitation vitamin A spectra with leave one-out cross-validation of: (a) Comisana with pasture feeding (•), Sicilo-Sarde with pasture feeding (º), Sicilo-Sarde feeding on scotch bean (▪), and Sicilo-Sarde feeding on soybean (□) groups; and (b) Sicilo-Sarde with pasture feeding (º), Sicilo-Sarde feeding on scotch bean (▪), and Sicilo-Sarde feeding on soybean (□) groups.
Global analysis of the milk samples based on excitation and emission fluorescence spectra throughout the lactation stage: concatenation
Milk is a complex product, which contains many intrinsic molecules that could fluoresce at specific excitation-emission wavelengths. The five fluorescence spectra acquired on milk samples from Sicilo-Sarde and Comisana ewes could be complementary. Taking this into consideration, a better discrimination between the investigated milk samples according to both feeding system and genotype could be found by jointly analysing the five spectral data sets. This process consists of putting the first ten PCs scores of the PCA applied on each data set in the same matrix, which is analysed by FDA. The similarity map of the FDA applied to the five fluorescence spectra is shown in . The four groups were well separated. Indeed: (i) Ssco milk samples presented positive scores according to discriminant factors 1 and 2; (ii) Spas group exhibited negative values according to discriminant factors 1 and 2; (iii) Ssoy milk samples had positive scores according to discriminant factor 1 and negative values according to discriminant factor 2; and (iv) Cpas group presented negative values according to discriminant factor 1 and positive scores following discriminant factor 2.
Figure 6 Discriminant analysis similarity map determined by discriminant factors 1 (F1) and 2 (F2). Factorial discriminant analysis (FDA) was performed on the concatenated PCs corresponding to the PCA performed on the emission spectra of aromatic amino acids and nucleic acids, tryptophan fluorescence spectra, riboflavin and vitamin A fluorescence spectra, and excitation spectra of vitamin A with leave one-out cross-validation of: (a) Comisana with pasture feeding (•), Sicilo-Sarde with pasture feeding (º), Sicilo-Sarde feeding on scotch bean (▪), and Sicilo-Sarde feeding on soybean (□) groups; and (b) Sicilo-Sarde with pasture feeding (º), Sicilo-Sarde feeding on scotch bean (▪), and Sicilo-Sarde feeding on soybean (□) groups.
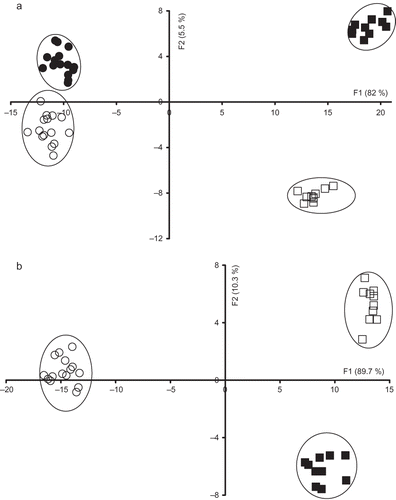
Correct classification amounting to 88% was observed following the application of FDA with leave one-out cross-validation. No milk sample belonging to Spas and Cpas groups was classified as Ssco or Ssoy groups and vice versa. The concatenation technique allowed also a good discrimination of milk samples according to the genotype, which was not observed when each fluorescence spectra was analysed separately, in agreement with Ereifej[Citation25] who succeeded to differentiate between three phenotype goat's milk during three lactation periods. Regarding Cpas group, only one sample was classified as belonging to Spas group, while three samples belonging to Spas group were assigned to Cpas group. Nine out of ten samples belonging to Ssoy and Ssco milk groups were correctly classified. The rate of correct classification was slightly lower to those found with Vis-NIR by Mouazen et al.[Citation26] although the latter authors did not apply FDA with leave one-out cross-validation. In addition, the similarity map gave a better separation of the investigated milk samples than with Vis-NIR.[Citation5]
The FDA applied to milk samples of the three Sicilo-Sarde ewes allowed also a good separation when milk samples originating from Sicilo-Sarde ewes were considered (). A correct classification rate amounting to 88.6% was observed, similar to that found with excitation and emission vitamin A spectra. Again, better differentiation was obtained with the concatenated fluorescence spectra than with Vis-NIR spectra.[Citation5]
CONCLUSION
The present investigation demonstrated the ability of front face fluorescence spectroscopy to monitor changes in ewe's milk according to both feeding system and genotype throughout the lactation period. By using, separately, each fluorescence spectra recorded after excitation set at 250, 290, 322, and 380 nm and emission set at 410 nm, the discrimination of the investigated milk samples were not satisfactory for differentiating the four groups. However, fluorescence spectroscopy demonstrated its ability to discriminate milk samples according to both the feeding system and genotype following the use of concatenation technique. Indeed, 88% of correct classification was obtained from the FDA, with leave one-out cross-validation applied to the four groups. The obtained results showed the robustness of the model obtained from ewe's milk samples collected from different: (i) genotype; (ii) feeding systems; (iii) ages; and (iv) lactation stage. Dairy plants in Tunisia are encouraged to adopt this technique coupled with multivariate statistical analyses to separate incoming milk from different farmers into various quality groups in order to preserve consistency of their final products. The development of chemometric tools and sensors allows us to foresee the usefulness of this technique as an on-line tool for the determination of the overall quality of complex food systems.
ACKNOWLEDGMENTS
The authors wish to acknowledge Dr. D. Bertrand (Inra Nantes, France) for “Saisir” software.
REFERENCES
- Casu , S. and Boyazoglu , J.G. 1990 . Mediterranean ovine milk production: Regions of production, genetic types used, breeding systems and future perspectives. . Options Méditerranéennes–Série A , 12 : 19 – 24 .
- Colson , F. , Kayouli , C. , Belloin , J.C. and Nardello , R. 1995 . Study of dairy cattle sector in Tunisia, TSS1-TUN/94/ 01T-001/AGAM-FAO
- Anonymous. Emergency Special Session of the General Assembly (ESSGA). United Nations. 2006 http://www.fao.org/es/ess/compendium_2006/pdf/TUN_ESS_E.pdf (http://http://www.fao.org/es/ess/compendium_2006/pdf/TUN_ESS_E.pdf)
- Schingoethe , J.D. , Rook , A.J. and Ludens , F. 1977 . Evaluation of sunflower meal as a protein supplement for lactating cows . Journal of Dairy Science , 60 : 591 – 595 .
- Mouazen , A.M. , Dridi , S. , Rouissi , H. , De Baerdemaeker , J. and Ramon , H. 2007 . Prediction of selected ewe's milk properties and differentiating between pasture and box feeding using visible and near infrared spectroscopy . Biosystems Engineering , 104 : 353 – 361 .
- Herbert , S. 1999 . Caractérisation de la structure moléculaire et microscopique de fromages à pâte molle. Analyse multivariée des données structurales en relation avec la texture , Nantes , France : Ecole Doctorale Chimie Biologie de l'Université de Nantes . PhD Thesis
- Herbert , S. , Riaublanc , A. , Bouchet , B. , Gallant , D.J. and Dufour , E. 1999 . Fluorescence spectroscopy investigations of acid-and rennet-induced milk coagulation of milk . Journal of Dairy Science , 82 : 2056 – 2062 .
- Karoui , R. , Martin , B. and Dufour , E. 2005 . Potentiality of front-face fluorescence spectroscopy to determine the geographic origin of milks from Haute-Loire department (France) . Le Lait , 85 : 223 – 236 .
- Hammami , M. , Rouissi , H. , Salah , N. , Selmi , H. , Al-Otaibi , M. , Blecker , C. and Karoui , R. 2010 . Fluorescence spectroscopy coupled with factorial discriminant analysis technique to identify sheep milk from different feeding systems . Food Chemistry , 122 : 1344 – 1350 .
- Maâmouri , O. , Rouissi , H. , Dridi , S. , Kammoun , M. , De Baerdemaeker , J. and Karoui , R. 2008 . Mid infrared attenuated total reflection spectroscopy as a rapid tool to assess the quality of Sicilo-Sarde ewe's milk during the lactation period after replacing soybean meal with scotch bean in the feed ration . Food Chemistry , 106 : 361 – 368 .
- Rouissi , H. , Dridi , S. , Kammoun , M. , De Baerdemaeker , J. and Karoui , R. 2008 . Front face fluorescence spectroscopy: A rapid tool for determining the effect of replacing soybean meal with scotch bean in the ration on the quality of Sicilo-Sarde ewe's milk during lactation period . European Food Research and Technology , 226 : 1021 – 1030 .
- Zaïdi , F. , Rouissi , H. , Dridi , S. , Kammoun , M. , De Baerdemaeker , J. and Karoui , R. 2008 . Front face fluorescence spectroscopy as a rapid and non destructive tool for differentiating between Sicilo-Sarde and Comisana ewe's milk during lactation period: A preliminary study . Food and Bioprocess Technology , 1 : 143 – 151 .
- Feinberg , M. , Dupont , D. , Efstathiou , T. , Louâore , V. and Guyonnet , J.P. 2006 . Evaluation of tracers for the authentification of thermal treatments of milks . Food Chemistry , 98 : 188 – 194 .
- Bertrand , D. and Scotter , C.N.G. 1992 . Application of multivariate analyses to NIR spectra of gelatinized starch . Applied Spectroscopy , 46 : 1420 – 1425 .
- Safar , M. , Bertrand , P.R. , Devaux , M.F. and Genot , C. 1994 . Characterization of edible oils, butters, and margarines by Fourier transform infrared spectroscopy with attenuated total reflectance . Journal of the American Chemical Society , 71 : 371 – 377 .
- Karoui , R. , Dufour , E. , Pillonel , L. , Picque , D. , Cattenoz , T. and Bosset , J.O. 2004 . Determining the geographic origin of Emmental cheeses produced during winter and summer using a technique based on the concatenation of MIR and fluorescence spectroscopic data . European Food Research and Technology , 219 : 184 – 189 .
- Miquel Becker , E. , Christensen , J. , Frederiksen , C.S. and Haugaard , V.K. 2003 . “ Front face fluorescence spectroscopy and chemometrics in analysis of yogurt: Rapid analysis of riboflavin ” . In Journal of Dairy Science86 2508 – 2515 .
- Dufour , E. and Riaublanc , A. 1997 . Potentiality of spectroscopic methods for the characterisation of dairy products, I—Front-face fluorescence study of raw, heated and homogenised milks . Le Lait , 77 : 657 – 670 .
- Karoui , R. , Dufour , E. , Pillonel , L. , Picque , D. , Cattenoz , T. and Bosset , J.O. 2004 . Fluorescence and infrared spectroscopies: A tool for the determination of the geographic origin of Emmental cheeses manufactured during summer . Le Lait , 84 : 359 – 374 .
- Karoui , R. , Dufour , E. , Pillonel , L. , Schaller , E. , Picque , D. , Cattenoz , T. and Bosset , J.O. 2005 . Determination of the geographic origin of Emmental cheeses by combining infrared and fluorescence spectroscopies . International Dairy Journal , 15 : 287 – 298 .
- Karoui , R. , Dufour , E. , Schoonheydt , R. and De Baerdemaeker , J. 2007 . Characterisation of soft cheese by front face fluorescence spectroscopy coupled with chemometric tools: Effect of the manufacturing process and sampling zone . Food Chemistry , 100 : 632 – 642 .
- Karoui , R. , Kemps , B. , Bamelis , F. , De Ketelaere , B. , Mertens , K. , Schoonheydt , R. , Decuypere , E. and De Baerdemaeker , J. 2006 . Development of a rapid method based on front face fluorescence spectroscopy for the monitoring of egg freshness: 2—Evolution of yolk . European Food Research and Technology , 223 : 180 – 188 .
- Karoui , R. , Nicolaï , B. and De Baerdemaeker , J. 2008 . Monitoring the egg freshness during storage under modified atmosphere by fluorescence spectroscopy . Food and Bioprocess Technology , 1 : 346 – 356 .
- Karoui , R. , Schoonheydt , R. , Decuypere , E. , Nicolaï , B. and De Baerdemaeker , J. 2007 . “ Front face fluorescence spectroscopy as a tool for the assessment of egg freshness during storage at a temperature of 12.2°C and relative humidity of 87% ” . In Analytical Chimica Acta582 83 – 91 .
- Ereifej , K.I. 2006 . Characteristics of milk, cheddar-like and white cheese obtained from three goat phenotypes during three lactation periods . International Journal of Food Properties , 9 : 803 – 811 .
- Mouazen , A.M. , Dridi , S. , Rouissi , H. , De Baerdemaeker , J. and Ramon , H. 2007 . Feasibility study on using VIS-NIR spectroscopy coupled with factorial discriminant analysis technique to identify sheep milk from different genotypes and feeding systems . Journal of Near Infrared Spectroscopy , 15 : 359 – 369 .