?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
This study presents an ecologically conscious methodology for synthesizing tea polypeptides-selenium nanoparticles (TP-SeNPs) exhibiting antioxidant and antibacterial properties, utilizing tea polypeptides (TP). Proteins were extracted from tea leaves using an alkali extraction-acid precipitation method, and response surface methodology (RSM) was employed to optimize the preparation processes for both TP and TP-SeNPs. The TP extract was obtained with the highest TP content of 28.12% at a pH of 6, temperature of 50°C, duration of 3 h, and enzyme concentration of 4%. TP-SeNPs with a minimum particle size of 98 nm were biosynthesized by adding 3% TP at pH 4 and reaction temperature of 55°C for 6 h. The UV spectrum of TP-SeNPs showed an absorption peak at 285 nm. The EDS peak at 1.37 keV confirmed the presence of selenium in TP-SeNPs. Electron microscopy revealed a thin layer coating on the surface of TP-SeNPs. Infrared analysis showed changes in the C=O, NH, and C-N bonds. XRD analysis indicated the formation of a crystalline structure in TP-SeNPs. The absorbance ratio of the peptide selenium nanoparticle colloidal solution remained stable over 7 days at 25°C and 4°C. TP-SeNPs exhibited higher antioxidant activity than SeNPs at high concentrations. TP-SeNPs contain selenium, carbon, oxygen, and nitrogen, and TP plays a role in stabilizing and encapsulating TP-SeNPs. This may involve physical adsorption, covalent interactions, and other unknown interactions, preventing aggregation or degradation. The crystalline structure of TP-SeNPs improves their material properties and enhances their antioxidant and antibacterial abilities.
Introduction
Selenium, a trace element, plays a pivotal role in numerous physiological processes, including immune system modulation, cardiovascular regulation, thyroid function maintenance, and anti-cancer properties.[Citation1] It is extensively utilized as a nutritional supplement in both inorganic forms, such as selenite and selenate, and organic forms, such as selenomethionine (SeMet) and selenocysteine (Sec).[Citation2] In comparison to inorganic and organic selenium, zero-valent oxidized SeNPs exhibit higher biological activity and lower toxicity.[Citation3]
In recent years, extensive investigations have been conducted physical and chemical methods for synthesizing SeNPs. However, there is a growing interest in the green synthesis of SeNPs using plant extracts, as they offer dual functionality as reducing agents and stabilizers.[Citation4,Citation5] Nonetheless, limited less research has been carried out on the synthesis and enhancement of biological activity of SeNPs using polypeptides. Tea, renowned for its therapeutic and nutritional advantages,[Citation6–8] has emerged as a subject of interest for scientists. Dried green tea contains approximately 28% proteins, and the hydrolysis of crude protein can lead to the production of active polypeptides with potential benefits.[Citation9] Proteolytic enzymes facilitate the catalysis of tea protein, resulting in the release of bioactive peptides that possess diverse physiological functions, including antioxidant, antibacterial, and immune regulation effects.[Citation10]
In this study, TP were obtained through enzymatic hydrolysis and employed for the green synthesis of TP-SeNPs using ascorbic acid and sodium selenite. The size, morphology, and binding mechanism of TP-SeNPs were characterized, and their stability and antioxidant properties were investigated and analyzed. The objective of this research was to produce biocompatible SeNPs composites mediated by plant-mediated TP, which hold promise for various biomedical applications.
Materials and method
Materials and reagents
Tea leaves were collected from the Donggyu tea garden located in Shaanxi, China and subjected to freeze-drying using a vacuum freeze dryer (LGJ-40E, Sihuan, Beijing, China). The freeze-dried tea leaves were then passed through a 60-mesh screen for further processing. Papain enzyme was procured from Shanghai Yuanye Biotechnology Co., Ltd. DPPH and ABTS were obtained from Fuyang Manlin Biotechnology Co., Ltd. Ascorbic acid was purchased from Shanghai Aladdin Biochemical Technology Co., Ltd. All other chemicals and reagents used in this study were of analytical grade.
Tea protein extraction
10 g of tea leaves were placed in a 250 mL NaOH solution with a pH of 10. The ultrasonic extraction (SB-5200DT, Xinzhi, Ningbo, China) was performed at 50°C for 100 min. Subsequently, the mixture was centrifuged (LC-800, Zhongjia, Hefei, China) at 4500 r/min for 15 min, and the resulting supernatant was collected. The residue underwent a second round of extraction, and the supernatants from both extractions were combined. The pH of the combined supernatants was adjusted to 4.2 using a 1 mol/L HCl solution. After pH adjustment, the mixture was allowed to settle for 12 h, followed by centrifugation at 6000 r/min for 20 min. The precipitate was collected and washed with distilled water until neutral. Finally, the collected precipitate was subjected to vacuum freeze-drying to obtain tea protein for future use.
Optimization of preparation process
Optimization of TP preparation Process: A 10 mg/mL tea protein solution was prepared by dissolving tea protein in 100 mL of distilled water. The effects of different extraction temperatures (40, 45, 50, 55, and 60°C), enzyme concentrations (1%, 2%, 3%, 4%, and 5% papain), solution pH values (5, 6, 7, 8, and 9), and extraction times (3, 6, 9, 12, and 15 h) were investigated to assess their influence on the yield of TP. Following the completion of the reaction, the solution’s pH was adjusted to neutral, and the mixture was subjected to heat treatment at 90°C for 15 min to inactivate the enzyme. The supernatant was then separated through centrifugation, and the tea protein was obtained by freeze-drying the collected supernatant.
In order to optimize the preparation process of TP, a Box-Behnken design (BBD) was employed to investigate the effects of independent variables and their respective levels () on the response of TP content. By analyzing the experimental data, the optimal parameters for the enzymatic hydrolysis process of tea protein were determined, and the response model was evaluated.
Table 1. Independent variables and levels used in Box-Behnken design for TP preparation.
Synthesis of SeNPs and optimization of TP-SeNPs preparation process: Initially, 200 mL of a 1 mM Na2SeO3 solution was gradually introduced to 200 mL of a 4 mmol/L Vc solution under continuous magnetic stirring, ensuring meticulous homogeneity. The pH of the resulting solution was adjusted to 6. Subsequently, the reaction was conducted at 55°C for 6 h. Upon completion, the reaction mixture was allowed to naturally equilibrate at ambient temperature. To ensure purification of the obtained SeNPs, the mixture was dialyzed with deionized water for 72 h, followed by concentration at 50°C and vacuum freeze-drying.
TP-SeNPs preparation was conducted following the methodology of previous studies with some modifications.[Citation11,Citation12] First, TP power (10, 20, 30, 40, 50 mg) was dissolved in 10 mL of distilled water. The TP solution was then mixed with 40 mL of a 1 mM Na2SeO3 solution and 40 mL of a 4 mM ascorbic acid solution. The pH of the solution was adjusted (2, 3, 4, 5, and 6), and the reaction was conducted at a fixed temperature (25, 35, 45, 55, and 65°C) and for the reaction time (3, 6, 9, 12, and 15 h). The influence of pH, temperature, time, and peptide solution concentration on the size of TP-SeNPs were investigated. TP-SeNPs were obtained by dialyzing with deionized water for 72 h, concentrating at 50°C, and then freeze-drying.
The BBD was employed to analyze the effect of the independent variables and their response levels on the particle size of TP-SeNPs (y) (). Based on the experimental data obtained from the response surface analysis, the optimal preparation parameters for TP-SeNPs were determined, and the response model was evaluated.
Table 2. Independent variables and levels used in Box-Behnken design for TP-SeNPs preparation.
Index determination
TP Content Determination: The TP content was determined using the TCA-N method,[Citation13] A 1.0 mg/mL TP solution was mixed with 20% TCA in a 1:1 volume ratio, followed by centrifugation at 4°C and 6000 rpm for 15 min. The nitrogen content in the supernatant was determined using the Kjeldahl method.
Particle Size Determination: Particle size was measured using a particle size analyzer (Nano ZS90, Malvern Instruments Ltd., UK). A 1 mg/mL solution of the sample was prepared, and three scans were performed with a single scan time of 13 s and 15 repeats per scan.
Characterization of TP, SeNPs and TP-SeNPs
A UV-Vis spectrophotometer (3900 U, Hitachi, Tokyo, Japan) was used to detect the optical characteristics of TP, SeNPs, and TP-SeNPs in the wavelength range of 200–800 nm. The surface morphology of TP, SeNPs, and TP-SeNPs was analyzed using a scanning electron microscope (Verios 460, Thermo Fisher, Waltham, US). The samples were placed on a clean silicon wafer and analyzed with an accelerating voltage of 15 kV. Elemental analysis of TP-SeNPs was conducted using EDX (Energy-dispersive X-ray spectroscopy). SAED (Selected Area Electron Diffraction) analysis was performed using a transmission electron microscope (JEM-2100F, Jeol, Tokyo, Japan).
FT-IR analysis was employed to determine the functional groups present in the samples by observing the absorption peaks and wavelengths in the infrared spectrum.[Citation14] 1 mg of the samples was mixed with 100 mg of KBr powder and prepared into tablets at 20MPa for 0.5 min. The spectral range analyzed was 4000–400 cm−1 using a spectrometer (vertex 70, Bruker, Karlsruhe, Germany).
XRD was performed using a Cu Kα radiation source (6100, Shimadzu, Kyoto, Japan) with an operating voltage of 40 kV and a current of 30 mA. The analysis encompassed a wide range of Bragg angles (5° ≤ 2θ ≤ 90°). The scanning speed was set to 12°/min, covering a range of 5–90° with a step size of 0.0131303.
Stability assessment
The TP-SeNPs and peptide-coated TP-SeNPs solutions were placed in glass vials and stored at 25°C for 7 days. The presence of any visible precipitate within the samples was visually assessed as an indicator of their stability. The absorbance values of the colloidal solutions were measured using a dual-wavelength spectrophotometric method.[Citation15] The colloidal particle size parameter B was calculated using the following formula:
where A500 and A420 represent the absorbance values at 500 nm and 420 nm, respectively. A value of B approaching 1 signifies a relatively stable colloidal particle size.
Antioxidant activity
DPPH Radical Scavenging activity: The DPPH radical scavenging activity was determined using a modified method as described by Baliyan.[Citation16] A 2 mL sample solution was mixed with 1 mL of 0.1 mM DPPH ethanol solution and incubated at 37°C in the dark for 30 min. The absorbance at 517 nm was measured. The DPPH radical scavenging activity was calculated using the following formula:
where As, Ac, and A0 represent the absorbance of the sample, control, and blank group, respectively.
ABTS Radical Scavenging Activity: The ABTS assay was performed following the method described by Wang.[Citation17] The ABTS solution (7.4 mM) and potassium persulfate solution (2.6 mM) were mixed in equal volume and allowed to react at room temperature in the dark for 16 h. The resulting solution was then diluted with 70% ethanol to obtain an absorbance of approximately 0.75 at 734 nm. To perform the assay, 600 µL of the ABTS solution was mixed with 25 µL of sample solution. After incubation for 10 min at room temperature, the absorbance at 734 nm was measured. The ABTS radical scavenging activity was calculated using the following formula:
where As, Ac, and A0 represent the absorbance of the sample, control, and blank group, respectively.
Hydroxyl Radical Scavenging Activity: The hydroxyl radical scavenging activity was assessed based on the Fenton reaction.[Citation18] A 1 mL sample solution was mixed with 1 mL of 9 mM FeSO4 solution, followed by the addition of 1 mL of a 9 mM salicylic acid solution, ensuring thorough mixing. Subsequently, 1 mL of 9 mM H2O2 was added to initiate the reaction. The mixture was then heated at 37°C in a light-protected water bath for 1 h. Afterward the absorbance of the mixture was measured at 510 nm. The hydroxyl radical scavenging activity was evaluated using the following formula:
where As, Ac, and A0 represent the absorbance of the sample, control, and blank groups, respectively.
FRAP: The FRAP assay was performed following the method developed by Sun.[Citation19] Briefly, a mixture of 1 mL of sample solution, 1 mL of 0.2 M sodium phosphate buffer (pH 7.2), and 1 mL of 1% K3[Fe(CN)6] was incubated at 50°C for 30 min. The resulting solution was then mixed with 1 mL of 10% trichloroacetic acid and centrifuged at 3500 r/min for 10 min. The supernatant (2 mL) was blended with 2 mL of distilled water and 0.4 mL of 0.1% FeCl3. The absorbance at 700 nm was measured, and the FRAP value was evaluated using the following formula:
where As, Ac, and A0 represent the absorbance of the sample, control, and blank
Antibacterial activity assay
Escherichia coli and Bacillus subtilis were cultured in Luria-Bertani (LB) liquid medium and incubated with shaking at 37°C for 24 h. The antibacterial activity of the sample was determined using the agar diffusion method, following a modified protocol based on Gonelimali et al.,[Citation20] 200 μL of bacterial suspension was spread evenly on the surface of an LB agar plate. Next, sterile filter paper discs with a diameter of 6 mm were dipped into different concentrations of the sample solution (2%, 4%, 6%, 8%, 10%) and placed on the agar plate, ensuring good contact with the medium. The plates were then incubated at 37°C for 18 h, with sterile water used as a blank control. The antibacterial activity was evaluated based on the diameter of the inhibition zone.
Cell viability assay
Cell viability was assessed through the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay to evaluate the cytotoxicity of the nanoparticles.[Citation21] Briefly, 100 μL of 293T cells at a density of 1 × 10[Citation4] cell/mL were seeded in a flat-bottom 96-well plate and incubated at 37°C with 5% CO2 for 24 h. TP-SeNPs at concentrations of 0, 2, 4, 6, 8, and 10 mg/mL were prepared in cell culture medium and added to the plate, with 5 replicate wells for each concentration. After 24 h of incubation, 10 μL MTT reagent was added to each well and incubated for 4 h. Then 100 μL of DMSO solution was added to each well. Finally, the OD value was measured at 570 nm.
Statistical analysis
Each experimental condition was repeated three times, and the results were expressed as mean ± standard deviation. The Design-Expert® software (ver. 9, Stat-Ease Inc., Minneapolis, USA) was utilized for designing the response surface experiments, and analysis of variance (ANOVA) was performed to evaluate the quality of the fitted mathematical models. Data processing and image generation were employed using Origin 8.0 software (OriginLab Corporation, USA). Statistical comparison between the mean values were conducted using either unpaired Student’s t-tests or ANOVA. A p-values threshold of less than 0.05 indicated a statistically significant difference between the means.
Results and discussion
Response surface analysis and process optimization
Proteolytic hydrolysis of proteins is a complex reaction process that is influenced by various factors, such as the type and properties of the substrate and enzyme, as well as temperature, pH value, hydrolysis time, and water volume.[Citation22] Therefore, RSM is employed to investigate the preparation process of TP.
The impact of four operational parameters (pH, temperature, time, and enzyme) on the TP content during the TP preparation process is presented in . A total of 27 experiments were conducted, resulting in peptide contents ranging from 8.32% to 28.12%. Sample 24 (pH 8, 40°C, 9 h, 4% papain) exhibited the lowest TP content of 8.32%, while sample 18 (pH 6, 50°C, 3 h, 4% papain) showed the highest content of 28.12%. These findings outperform the hydrolysis method utilizing pancreatic protease in black tea.[Citation23] Based on the experimental results, a quadratic function model was fitted as follows:
Table 3. Response Surface Design and Results of the Tea Polypeptides Preparation.
where Y represents the predicted value of the response variable. A corresponds to the pH value, B corresponds to the temperature, C corresponds to the reaction time, and D corresponds to the enzyme concentration. The positive or negative sign preceding each coefficient signifies the positive or negative impact of the respective factor on the response variable. The coefficient of determination (R2) was found to be 0.977, indicating a strong fit between the model and the observed data.
The ANOVA analysis (Table S1) revealed that the regression model for TP preparation process was significant (P < .0001). Factors B and D, as well as the quadratic terms A,[Citation2] B,[Citation2] C,[Citation2] and D,[Citation2] exhibited significant effects on peptide preparation (P < .05). Additionally, the interaction terms AB and CD had significant impacts on the response variable (P < .05). This suggests that there are interactive effects between pH value and temperature, as well as reaction time and enzyme concentration, influencing the content of tea peptides.
Enzymatic hydrolysis is a preferred method in the food industry due to its mild process conditions, high specificity, and minimal byproduct formation.[Citation24] To enhance the efficiency of enzymatic hydrolysis, optimization can be performed in various aspects. However, long hydrolysis times, low analyte recovery, and the high solid content effect can potentially impact the accuracy of the results.[Citation25] To determine the optimal synthesis conditions, RSM has been used to establish an empirical model.[Citation26] presents the effects of pH, temperature, time, and TP concentration on the particle size of TP-SeNPs, including 27 experimental runs.
Table 4. Response Surface Design and Results of the Tea peptide selenium nanoparticles Process.
The particle size of TP-SeNPs ranged from 98 nm to 218 nm, depending on the conditions utilized in each experimental run. Experiment 7, with pH 4, 55°C, 6 h, and 3 mg/mL TP concentration, produced the minimum particle size of 98 nm. Conversely, experiment 9, with a pH of 5, temperature of 45°C, time of 6 h, and 5 mg/mL TP concentration, yielded the maximum particle size of 218 nm. The remaining experimental runs presented in the table also manifested varied particle sizes, which accentuates the impact of each factor on the size of TP-SeNPs. The fitting equation of TP-SeNPs was found to be:
The coefficient of determination (R2) of 0.965 indicates that the model is suitable for explaining the relationship between the factors and the response variable. The ANOVA analysis (Table S2) indicates that the model has a significant effect in predicting particle size (p < .001). The p-value for the lack-of-fit test is 0.769, suggesting that the model is suitable for predicting particle size.
The factors B (temperature), D (TP concentration), and AC (interaction between pH and time) have the most significant impacted on the particle size of TP-SeNPs (p < .001). The factors A/AB and AD/BD also have a significant effect on TP-SeNPs particle size (p < .05).
Figure S1 illustrates the influence of the interaction among factors on the particle size of TP-SeNPs. With an increase in reaction temperature and pH, the size of peptide selenium nanoparticles gradually increases. With an increase in reaction time and pH, the particle size of selenium nanoparticles initially increases and then decreases. With an increase in peptide concentration and pH, the particle size of selenium nanoparticles shows a gradual increasing trend. As the peptide concentration increases, the particle size also increases, while with an increase in temperature, the particle size initially increases and then decreases.
The optimal conditions for minimizing TP-SeNPs particle size were identified by employing the function method, resulting in a pH of 3.94, extraction temperature of 35.29°C, reaction time of 8.79 h, and TP concentration of 3.2 mg/mL. Under these conditions, the TP-SeNP particle size obtained was 91.48 nm. This is consistent with the particle size of soy peptide-chelated selenium, which ranges from 190.1 to 91.28 nm.[Citation27]
Characterization of TP, SeNPs and TP-SeNPs
The characterization was performed using UV, EDS, SEM, FT-IR, and XRD. As shown in , the UV spectrum of SeNPs exhibits a broad peak around 285 nm, which is consistent with the results of chemical and green synthesis of SeNPs.[Citation28] The absorption peak of TP-SeNPs exhibits a redshift, with the maximum peak appearing at 307 nm. This shift is likely attributed to the interaction between peptides and selenium nanoparticles, leading to changes in the electron resonance structure and subsequently affecting the position of the absorption peak.
Figure 1. The characterization of TP, SeNPs, and TP-SeNPs. (a) UV spectrum, (b) EDS, (c) SEM image of TP, (d) SEM images of SeNPs, (e) TEM image of TP-SeNPs ,(f) SEM image of TP-SeNPs, (g) FT-IR spectra, (h) XRD spectra.
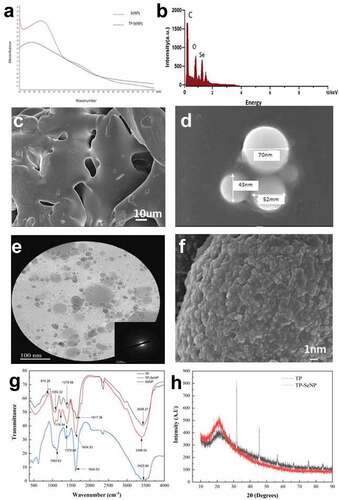
EDS shown in exhibits a peak at 1.37 keV, indicating the presence of Se and confirming the biological reduction of selenium ions into elemental form. This finding is consistent with the results reported by D. Cui.[Citation29] The analysis also reveals the presence of carbon, followed by oxygen and nitrogen, in the TP-SeNPs. These elements are derived from the tea peptides that are bound to the surface of SeNPs.
The surface morphology and microstructure of TP, SeNPs, and TP-SeNPs were visualized and compared in . As displayed in , TP exhibits a sleek sheet structure. In , spherical-shaped SeNPs are observed, with a particle size ranging from 40–70 nm, and these nanoparticles tend to aggregate. shows the SAED pattern, indicating that TP-SeNPs exhibit a locally ordered polycrystalline structure. Additionally, reveals a thin layer enveloping the surface of TP-SeNPs, which is presumed to be the coating of TP. This coating plays a crucial role in stabilizing the nanoparticles and preventing aggregation, as observed in a previous studies.[Citation11,Citation30] Therefore, this phenomenon suggests that TP acts as a stabilizer, effectively interacting with the surface of SeNPs and forming a protective layer that hinders particle-particle interactions and aggregation.
To further investigate the interaction mechanism between TP and stabilized SeNPs, FT-IR analysis was conducted on TP, SeNPs, and TP-SeNPs (). A blue shift peak at 3398.04 cm−1 is observed in TP-SeNPs. This shift could be attributed to the -OH and -NH functional groups in TP. It suggests that the -OH and -NH groups in TP may form hydrogen bond interactions with SeNPs, stabilizing the TP-SeNPs through physical adsorption. This interaction is likely to reduce the aggregation and degradation of SeNPs and enhance the stability of the complex. Meanwhile, the peaks of -C=O bond derived from TP at 1604.53 cm−1 and 1375.69 cm−1 show significantly shifts, suggesting that the -C=O group may be the binding site of SeNPs. This suggests that the -C=O functional groups in TP may interact with SeNPs through coordination and interact with the surface of SeNPs.TP contains amino acid residues, and the specific shift or intensity change of the amine (NH) vibration band at 1050.32 cm-1 in TP-SeNPs suggests that the C-N bonds in TP may interact with SeNPs, leading to changes of the C-N bonds. Based on the evidence presented, it can be inferred that the stabilization of SeNPs by TP may involve physical adsorption, covalent interaction, and other unknown interactions, which help prevent the aggregation or degradation of the complex.[Citation31,Citation32]
To determine the crystal structure of TP-SeNPs, XRD analysis was performed on both TP and TP-SeNPs (). When a crystalline material is exposed to X-rays, it produces sharp, distinct peaks, while amorphous materials exhibit broad hump peaks. The XRD spectrum of SeNPs does not exhibit any sharp diffracti[Citation15]on peaks, indicating that they are amorphous. In contrast, the XRD of TP-SeNPs displays a strong peak, suggesting that the TP-SeNPs formed by TP possess a crystalline structure.[Citation33]
The transition from an amorphous to a crystalline structure during synthesis involves a shift from an irregular to a well-defined, and ordered internal structure with flat surfaces. This can lead to improved material properties, such as increased stability, higher melting points, and enhanced optical or electrical properties, compared to amorphous materials with a disordered structure.[Citation34]
Stability
The stability of SeNPs and TP-SeNPs was investigated under pH 8 at 4°C and 25°C (). The results showed that the SeNPs solution exhibited a red sediment at the bottom after being stored for 7 days (). In contrast, the TP-SeNPs solution remained stable without any precipitation (). The colloidal particle size parameters were calculated by measuring the absorbance of the solutions (). The absorbance ratio of TP-SeNPs solution remained relatively stable under both room temperature and 4°C conditions, while the absorbance of SeNPs solution showed an increasing trend. This indicates that TP plays a positive role in enhancing the stabilizing of SeNPs, preventing their aggregation and precipitation. This is crucial for the long-term stability and controllability of SeNPs in practical applications.
Antioxidant activity
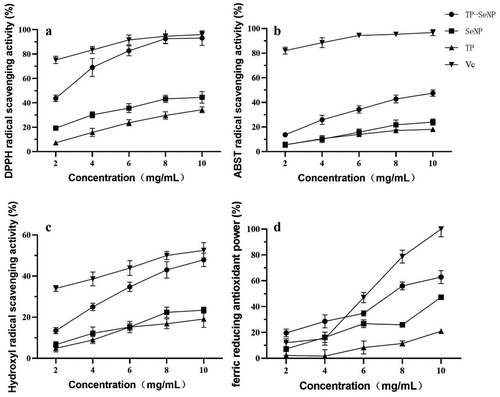
demonstrates the performance of the samples in terms of DPPH, ABTS, FRAP, and hydroxyl radical scavenging activities.
presents the DPPH radical scavenging activity of TP-SeNP, SeNP, and TP at various concentrations (mg/mL). As depicted, the samples displayed a dose-response correlation. The order of decreasing DPPH radical scavenging activity among the samples was as follows: Vc (IC50 = 0.79 mg/mL) > TP-SeNPs (IC50 = 2.37 mg/mL) > SeNPs (IC50 = 12.47 mg/mL) > TP (IC50 = 17.21 mg/mL). Menon et al.[Citation35] synthesized SeNPs using ginger extract, with nanoparticle sizes ranging from 100–150 nm, similar to TP-SeNPs. The DPPH assay revealed an IC50 of 125 μg/mL, which outperformed TP-SeNPs.[Citation36] shows the ABTS radical scavenging activity of TP, SeNPs, and TP-SeNPs at different concentrations. The results indicate that as the concentration of the samples increased, their ABTS radical scavenging activity increased. TP-SeNPs showed the highest ABTS radical scavenging activity among all samples at all concentration levels. The order of decreasing ABTS radical scavenging activity among the samples was as follows: Vc (IC50 = 8.72 mg/mL) > TP-SeNPs (IC50 = 10.56 mg/mL) > SeNPs (IC50 = 34.16 mg/mL) > TP (IC50 = 46.22 mg/mL). Overall, TP-SeNPs has better antioxidant and reducing capacity than TP and SeNPs, but its activity is lower than that of Vc. This is comparable to the ABTS radical scavenging activity of SeNPs functionalized with prickly pear polysaccharides, which was reported to be around 50%.[Citation37] The results presented in demonstrate the OH radical scavenging ability of TP-SeNP, SeNP, and TP. As the concentration of the samples increased, their ability to scavenge OH radicals increased.[Citation38] The IC50 for OH radical scavenging activity of TP-SeNP was 10.56 mg/mL. The order of decreasing OH radical scavenging activity among the samples was: TP-SeNP > SeNP > TP, with TP-SeNP exhibiting the highest activity of 46.5% at a concentration of 10 mg/mL. However, this activity was lower compared to soy peptide-chelated selenium at a concentration of 1 mg/mL, which showed a scavenging rate of 61%.[Citation27] illustrates the FRAD of TP, SeNPs, and TP-SeNPs at different concentrations. As the concentration of the samples increased, their antioxidant power also increased. TP-SeNPs exhibited the highest antioxidant power, with a maximum reduction power of 62.9% at a concentration of 10%. Overall, TP-SeNPs had better antioxidant power than TP and SeNPs, but their activity was still lower than Vc. Additionally, selenium nanoparticles mediated by Diospyros montana leaf extract (4–16 nm) exhibited better reduction power than standard ascorbic acid.[Citation39]
By utilizing multiple assay methods including DPPH, ABTS, and FRAP, the antioxidant behavior of TP-SeNPs was comprehensively analyzed, revealing their antioxidant potential from different perspectives. This comprehensive evaluation also helps to compensate for the discrepancies that may exist among individual methods in terms of antioxidant mechanisms, redox potentials, substrate types, chemical conditions, and presentation of results.[Citation40,Citation41]
SeNPs have antioxidant properties and can eliminate free radicals in the body. This characteristic is attributed to their small size, large surface area, and unique electronic properties, with smaller nanoparticles exhibiting stronger antioxidant activity.[Citation35] Peptides, on the other hand, contain amino acids and also possess antioxidant properties due to their ability to scavenge free radicals and chelate metal ions.[Citation42] The combination of peptides and SeNPs may result in a synergistic antioxidant effect. This is because peptides can stabilize SeNPs and enhance their antioxidant performance.[Citation43] Additionally, studies have shown that the antioxidant mechanism of selenopeptides involves the regulation of the Keap1-Nrf2 pathway, which plays a crucial role in cellular defense against oxidative stress.[Citation44]
Antibacterial activity
shows the antibacterial activity of TP, SeNPs, and TP-SeNPs against Escherichia coli and Bacillus subtilis at different concentrations. TP-SeNPs exhibited a significantly larger bacteriostatic circle compared to TP and SeNPs against Escherichia coli at 2%, 4%, and 6% concentration, as well as against Bacillus subtilis at 8% and 10% concentration. TP-SeNPs had a higher antibacterial activity against Escherichia coli than Bacillus subtilis, and the antibacterial activity improved as the concentration increased. The largest inhibition zone was observed for 10% TP-SeNPs, indicating that TP-SeNPs have the highest antibacterial activity.
Table 5. Inhibitory effects of TP, SeNPs and TP-SeNPs against Escherichia coli and Bacillus subtilis.
The antibacterial activity of SeNPs is influenced by factors such as concentration, size, and synthesis method.[Citation45] Within the range of 81–205 nm, the antibacterial performance of SeNPs decreases as their particle size increases.[Citation46] Oxidative damage may play an important role in the antibacterial process, and the addition of functional ingredients to SeNPs can enhance their antibacterial activity.[Citation47]
SeNPs possess antibacterial properties, possibly due to their interaction with bacterial cell membranes, leading to membrane damage and cell death.[Citation48] SeNPs can also enhance the antibacterial activity of peptides by providing additional membrane-disrupting agents.[Citation43] The antibacterial ability of SeNPs is influenced by concentration, size, and synthesis methods.[Citation45] Within the range of 81–205 nm, the antibacterial performance of SeNPs decreases with increasing particle size.[Citation49] Peptides also exhibit antibacterial properties against Pseudomonas aeruginosa, possibly through their ability to disrupt bacterial membranes and inhibit bacterial growth.[Citation50] The combination of peptides and SeNPs may result in a synergistic antibacterial effect, where peptides enhance the antibacterial performance of SeNPs by promoting their interaction with bacterial cells and increasing uptake. Additionally, oxidative damage may play an important role in the antibacterial process, and the addition of functional components to SeNPs can enhance their antibacterial activity.[Citation47]
Cell viability
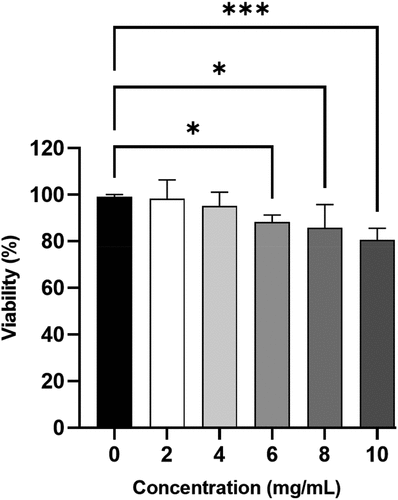
The effect of TP-SeNPs on the cell viability of 293T cells was determined using the MTT assay. As shown in , the cell viability of TP-SeNPs significantly decreased with increasing concentration. When TP-SeNPs reached 6 mg/mL, a decrease in cell viability was observed, and at 10 mg/mL, the cell viability was measured to be 80.6%. In comparison to ZnO-NPs and ZnO-PEG-NPs synthesized by Ebadi, which maintained a cell viability above 90% at a concentration of 75 mg/mL, TP-SeNPs exhibited higher cytotoxicity.[Citation51] SeNPs exhibited a concentration-dependent induction of cell apoptosis. SeNPs at concentrations below 172 μg/mL did not show cytotoxic effects on A-172, Caco-2, DU-145, and MCF-7 cell lines. However, SeNPs enhanced the expression of pro-apoptotic genes in almost all cancer cell lines except Caco-2, and activated various pathways involved in the adaptive unfolded protein response and pro-apoptotic signaling pathways.[Citation52]
Conclusion
In comparison to microbial agents, the utilization of plant extracts in green synthesis has gained popularity due to its eco-friendliness, use of biodegradable reducing agents, and cost reduction. In this study, tea protein was obtained from tea leaves using an alkaline extraction-acid precipitation method. TP extract with a content of 28.12% was obtained by hydrolyzing tea protein at pH 6, temperature 50°C, for 3 h, with a concentration of 4% papain. TP-SeNPs with a minimum particle size of 98 nm were biologically synthesized by adding 3% TP at pH 4 and a reaction temperature of 55°C for 6 h. TP-SeNPs exhibited absorption peaks of SeNPs at 285 nm in the UV spectrum, and the presence of selenium was confirmed by the peak at 1.37 keV in EDS, along with carbon, oxygen, and nitrogen, originating from tea peptides bound to the surface of selenium nanoparticles. Electron microscopy showed a thin layer coating on the surface of TP-SeNPs, indicating the stabilizing effect of TP, ensuring the stability and preventing aggregation of the nanoparticles. Infrared spectroscopy revealed changes in C=O, NH, and C-N, suggesting that TP’s stabilization of SeNPs may involve physical adsorption, covalent interactions, and other unknown interactions, preventing the aggregation or degradation of the complex. XRD analysis demonstrated the crystalline structure of TP-SeNPs, indicating a transition from amorphous to crystalline structure, which may enhance the material properties and improve the antioxidant and antibacterial abilities of TP-SeNPs. Over a period of 7 days at 25°C and 4°C, the absorbance ratio of the peptide-nano selenium colloid solution remained stable. TP-SeNPs exhibited higher antioxidant activity than SeNPs at high concentrations, and 10% TP-SeNPs showed good antibacterial activity. This study presents a novel and innovative approach to synthesizing SeNPs using plant-derived polypeptides, with potential applications in various fields, including the food and pharmaceutical industries. Further research is needed to evaluate the biocompatibility and safety of TP-SeNPs, including cytotoxicity and biocompatibility tests. Additionally, determining the optimal size range for the antioxidant and antibacterial properties of TP-SeNPs requires further investigation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Zhang, X.; He, H.; Xiang, J.; Yin, H.; Hou, T. Selenium-Containing Proteins/Peptides from Plants: A Review on the Structures and Functions. J. Agric. Food Chem. 2020, 68(51), 15061–15073. DOI: 10.1021/acs.jafc.0c05594.
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium Nanoparticles as a Nutritional Supplement. Nutrition. 2017, 33, 83–90. DOI: 10.1016/j.nut.2016.05.001.
- Benko, I.; Nagy, G.; Tanczos, B.; Ungvari, E.; Sztrik, A.; Eszenyi, P.; Prokisch, J.; Banfalvi, G. Subacute Toxicity of Nano-Selenium Compared to Other Selenium Species in Mice. Environ. Toxicol. Chem. 2012, 31(12), 2812–2820. DOI: 10.1002/etc.1995.
- Fardsadegh, B.; Jafarizadeh-Malmiri, H. Aloe Vera Leaf Extract Mediated Green Synthesis of Selenium Nanoparticles and Assessment of Their in vitro Antimicrobial Activity Against Spoilage Fungi and Pathogenic Bacteria Strains. Green Process. Synth. 2019, 8(1), 399–407. DOI: 10.1515/gps-2019-0007.
- Yilmaz, M. T.; Ispirli, H.; Taylan, O.; Dertli, E. A Green Nano-Biosynthesis of Selenium Nanoparticles with Tarragon Extract: Structural, Thermal, and Antimicrobial Characterization. LWT Food Sci. Technol. 2021, 141, 141. DOI: 10.1016/j.lwt.2021.110969.
- Klepacka, J.; Tońska, E.; Rafaowski, R.; Czarnowska-Kujawska, M.; Opara, B. Tea as a Source of Biologically Active Compounds in the Human Diet. Molecules. 2021, 26(5), 1487. DOI: 10.3390/molecules26051487.
- Xie, G.; Yan, J.; Lu, A.; Kun, J.; Wang, B.; Song, C.; Tong, H.; Meng, Q. Characterizing Relationship Between Chemicals and in vitro Bioactivities of Teas Made by Six Typical Processing Methods Using a Single Camellia Sinensis Cultivar, Meizhan. Bioengineered. 2021, 12(1), 1251–1263. DOI: 10.1080/21655979.2021.1903237.
- Koch, W.; Zagórska, J.; Marzec, Z.; Kukula-Koch, W. Applications of Tea (Camellia Sinensis) and Its Active Constituents in Cosmetics. Molecules. 2019, 24(23), 4277. DOI: 10.3390/molecules24234277.
- Cui, Q.; NI, X.; ZENG, L.; TU, Z.; LI, J.; SUN, K.; CHEN, X.; LI, X. Optimization of Protein Extraction and Decoloration Conditions for Tea Residues. Hortic. Plant J. 2017, 3(4), 172–176. DOI: 10.1016/j.hpj.2017.06.003.
- Xu, Y. Q.; Zhong, X. Y.; Chen, S. Q.; Yin, J. F. Hydrolysis of Green Tea Residue Protein Using Proteolytic Enzyme Derived from Aspergillus Oryzae. J. Food Sci. Technol. 2011, 50(1), 171–175. DOI: 10.1007/s13197-011-0239-x.
- Ye, M. J.; Xu, Q. L.; Tang, H. Y.; Jiang, W. Y.; Su, D.-X.; He, S.; Zeng, Q.-Z.; Yuan, Y. Development and Stability of Novel Selenium Colloidal Particles Complex with Peanut Meal Peptides. LWT- Food Sci. Technol. 2020, 126, 109280. DOI: 10.1016/j.lwt.2020.109280.
- Sharma, S.; Baboota, S.; Amin, S.; Mir, S. R. Ameliorative Effect of a Standardized Polyherbal Combination in Methotrexate-Induced Nephrotoxicity in the Rat. Pharm. Biol. 2020, 58(1), 184–199. DOI: 10.1080/13880209.2020.1717549.
- Li, C.; Zhang, B.; Wang, X.; Pi, X.; Wang, X.; Zhou, H.; Mai, K.; He, G. Improved Utilization of Soybean Meal Through Fermentation with Commensal Shewanella sp. MR-7 in Turbot (Scophthalmus Maximus L.). Microb. Cell Fact. 2019, 18(1), 214. DOI: 10.1186/s12934-019-1265-z.
- Liu, Y.; Huang, W.; Han, W.; Li, C.; Zhang, Z.; Hu, B.; Chen, S.; Cui, P.; Luo, S.; Tang, Z., et al. Structure Characterization of Oudemansiella Radicata Polysaccharide and Preparation of Selenium Nanoparticles to Enhance the Antioxidant Activities - ScienceDirect. LWT. 2021, 146, 111469. DOI: 10.1016/j.lwt.2021.111469.
- Shard, A. G.; Wright, L.; Minelli, C. Robust and Accurate Measurements of Gold Nanoparticle Concentrations Using UV-Visible Spectrophotometry. Biointerphases. 2018, 13(6), 061002. DOI: 10.1116/1.5054780.
- Baliyan, S.; Mukherjee, R.; Priyadarshini, A.; Vibhuti, A.; Gupta, A.; Pandey, R. P.; Chang, C.-M. Determination of Antioxidants by DPPH Radical Scavenging Activity and Quantitative Phytochemical Analysis of Ficus Religiosa. Molecules. 2022, 27(4), 1326. DOI: 10.3390/molecules27041326.
- Wang, M.; Li, C.; Li, H.; Wu, Z.; Chen, B.; Lei, Y.; Shen, Y. In vitro and in silico Antioxidant Activity of Novel Peptides Prepared from Paeonia Ostii ‘Feng Dan’hydrolysate. Antioxidants. 2019, 8(10), 433. DOI: 10.3390/antiox8100433.
- Wang, R.; Zhao, H.; Pan, X.; Orfila, C.; Lu, W.; Ma, Y. Preparation of Bioactive Peptides with Antidiabetic, Antihypertensive, and Antioxidant Activities and Identification of α-Glucosidase Inhibitory Peptides from Soy Protein. Food Sci. Nutr. 2019, 7(5), 1848–1856. DOI: 10.1002/fsn3.1038.
- Sun, Y.; Li, S.; Zeng, F.; Qi, J.; Qin, W.; Tan, C.; Luo, Q.; Wu, D.; Zhang, Q.; Lin, D., et al. Functional Components, Antioxidant Activity and Hypoglycemic Ability Following Simulated Gastro-Intestinal Digestion of Pigments from Walnut Brown Shell and Green Husk. Antioxidants (Basel). 2019, 8(12), 573.
- Gonelimali, F. D.; Lin, J.; Miao, W.; Xuan, J.; Charles, F.; Chen, M.; Hatab, S. R. Antimicrobial Properties and Mechanism of Action of Some Plant Extracts Against Food Pathogens and Spoilage Microorganisms. Front. Microbiol. 2018, 9, 1–9. DOI: 10.3389/fmicb.2018.01639.
- Ghaffari, H.; Tavakoli, A.; Moradi, A.; Tabarraei, A.; Bokharaei-Salim, F.; Zahmatkeshan, M.; Farahmand, M.; Javanmard, D.; Kiani, S. J.; Esghaei, M., et al. Inhibition of H1N1 Influenza Virus Infection by Zinc Oxide Nanoparticles: Another Emerging Application of Nanomedicine. J. Biomed. Sci. 2019, 26(1), 70.
- Chew, L. Y.; Toh, G. T.; Ismail, A. Chapter 15 - Application of Proteases for the Production of Bioactive Peptides. In Enzymes in Food Biotechnology; Kuddus, M. Ed.;Academic Press:2019; pp. 247–261. DOI: 10.1016/B978-0-12-813280-7.00015-3
- Su, K.; Mao, X.; Ai, L.; Zhang, X. In vitro Assessment of Anti-Diabetic Potential of Four Kinds of Dark Tea (Camellia Sinensis L.) Protein Hydrolysates. J. Appl. Botany Food Qual. 2019, 92, 57–63.
- Cruz-Casas, D. E.; Aguilar, C. N.; Ascacio-Valdés, J. A.; Rodríguez-Herrera, R.; Chávez-González, M. L.; Flores-Gallegos, A. C. Enzymatic Hydrolysis and Microbial Fermentation: The Most Favorable Biotechnological Methods for the Release of Bioactive Peptides. Food Chem. Mole. Sci. 2021, 3, 100047. DOI: 10.1016/j.fochms.2021.100047.
- da Silva, A. S. A.; Espinheira, R. P.; Teixeira, R. S. S.; de Souza, M. F.; Ferreira-Leitão, V.; Bon, E. P. S. Constraints and Advances in High-Solids Enzymatic Hydrolysis of Lignocellulosic Biomass: A Critical Review. Biotechnol Biofuels. 2020, 13(1), 58. DOI: 10.1186/s13068-020-01697-w.
- El-Malah, Y.; Nazzal, S.; Khanfar, N. M. D-Optimal Mixture Design: Optimization of Ternary Matrix Blends for Controlled Zero-Order Drug Release from Oral Dosage Forms. Drug Dev. Indus. Pharm. 2008, 32(10), 1207–1218. DOI: 10.1080/03639040600685167.
- Ye, Q.; Wu, X.; Zhang, X.; Wang, S. Organic Selenium Derived from Chelation of Soybean Peptide-Selenium and Its Functional Properties in vitro and in vivo. Food Funct. 2019, 10(8), 4761–4770. DOI: 10.1039/C9FO00729F.
- Pillay, N. S.; Daniels, A.; Singh, M. Folate-Targeted Transgenic Activity of Dendrimer Functionalized Selenium Nanoparticles in vitro. Int. J. Mol. Sci. 2020, 21(19), 7177. DOI: 10.3390/ijms21197177.
- Cui, D.; Yan, C.; Miao, J.; Zhang, X.; Chen, J.; Sun, L.; Meng, L.; Liang, T.; Li, Q. Synthesis, Characterization and Antitumor Properties of Selenium Nanoparticles Coupling with Ferulic Acid. Mater. Sci. Eng. C. 2018, 90, 104–112. DOI: 10.1016/j.msec.2018.04.048.
- Pyrzynska, K.; Sentkowska, A. Biosynthesis of Selenium Nanoparticles Using Plant Extracts. J. Nanostruct. Chem. 2022, 12(4), 467–480. DOI: 10.1007/s40097-021-00435-4.
- Dugyala, V. R.; Muthukuru, J. S.; Mani, E.; Basavaraj, M. G. Role of Electrostatic Interactions in the Adsorption Kinetics of Nanoparticles at Fluid–Fluid Interfaces. Phys. Chem. Chem. Phys. 2016, 18(7), 5499–5508. DOI: 10.1039/C5CP05959C.
- Sahu, D.; Jana, K.; Ganguly, B. The Role of Non-Covalent Interaction for the Adsorption of CO2 and Hydrocarbons with Per-Hydroxylated Pillar[6]arene: A Computational Study. New. J. Chem. 2017, 41(20), 12044–12051. DOI: 10.1039/C7NJ01744H.
- Wu, Y.; Liu, H.; Li, Z.; Huang, D.; Nong, L.; Ning, Z.; Hu, Z.; Xu, C.; Yan, J.-K. Pectin-Decorated Selenium Nanoparticles as a Nanocarrier of Curcumin to Achieve Enhanced Physicochemical and Biological Properties. IET Nanobiotechnol. 2019, 13(8), 880–886. DOI: 10.1049/iet-nbt.2019.0144.
- Xiong, Z.; Sun, B.; Zou, H.; Wang, R.; Fang, Q.; Zhang, Z.; Qiu, S. Amorphous-To-Crystalline Transformation: General Synthesis of Hollow Structured Covalent Organic Frameworks with High Crystallinity. J. Am. Chem. Soc. 2022, 144(14), 6583–6593. DOI: 10.1021/jacs.2c02089.
- Menon, S.; KS, S. D.; Agarwal, H.; Shanmugam, V. K. Efficacy of Biogenic Selenium Nanoparticles from an Extract of Ginger Towards Evaluation on Anti-Microbial and Anti-Oxidant Activities. Colloid Interface Sci. Commun. 2019, 29, 1–8. DOI: 10.1016/j.colcom.2018.12.004.
- Kokila, K.; Elavarasan, N.; Sujatha, V. Diospyros Montana Leaf Extract-Mediated Synthesis of Selenium Nanoparticles and Their Biological Applications. New. J. Chem. 2017, 41(15), 7481–7490. DOI: 10.1039/C7NJ01124E.
- Chen, W.; Cheng, H.; Xia, W. Construction of Polygonatum Sibiricum Polysaccharide Functionalized Selenium Nanoparticles for the Enhancement of Stability and Antioxidant Activity. Antioxidants (Basel). 2022, 11(2), 240. DOI: 10.3390/antiox11020240.
- Apak, R.; Özyürek, M.; Güçlü, K.; Çapanoğlu, E. Antioxidant Activity/Capacity Measurement. 1. Classification, Physicochemical Principles, Mechanisms, and Electron Transfer (ET)-Based Assays. J. Agric. Food Chem. 2016, 64(5), 997–1027. DOI: 10.1021/acs.jafc.5b04739.
- Bharathi, D.; Diviya Josebin, M.; Vasantharaj, S.; Bhuvaneshwari, V. Biosynthesis of Silver Nanoparticles Using Stem Bark Extracts of Diospyros Montana and Their Antioxidant and Antibacterial Activities. J. Nanostruct. Chem. 2018, 8(1), 83–92. DOI: 10.1007/s40097-018-0256-7.
- Apak, R.; Gorinstein, S.; Böhm, V.; Schaich, K. M.; Özyürek, M.; Güçlü, K. Methods of Measurement and Evaluation of Natural Antioxidant Capacity/Activity (IUPAC Technical Report). Pure Appl. Chem. 2013, 85(5), 957–998. DOI: 10.1351/PAC-REP-12-07-15.
- Xiao, F.; Xu, T.; Lu, B.; Liu, R. Guidelines for Antioxidant Assays for Food Components. Food Front. 2020, 1(1), 60–69. DOI: 10.1002/fft2.10.
- Olszowy-Tomczyk, M. Synergistic, Antagonistic and Additive Antioxidant Effects in the Binary Mixtures. Phytochem Rev. 2020, 19(1), 63–103. DOI: 10.1007/s11101-019-09658-4.
- Bankier, C.; Matharu, R. K.; Cheong, Y. K.; Ren, G. G.; Cloutman-Green, E.; Ciric, L. Synergistic Antibacterial Effects of Metallic Nanoparticle Combinations. Sci. Rep. 2019, 9(1), 16074. DOI: 10.1038/s41598-019-52473-2.
- Ranjitha, V. R.; Rai, V. R. Selenium Nanostructure: Progress Towards Green Synthesis and Functionalization for Biomedicine. J. Pharm. Invest. 2021, 51(2), 117–135. DOI: 10.1007/s40005-020-00510-y.
- Hariharan, H.; Al-Dhabi, N. A.; Karuppiah, P.; Rajaram, S. K. Nanocomposite Using Saccharomyces cerevisiae and Its Antimicrobial Activity Against Pathogens Causing Nosocomial. Chalcogenide Letters. 2012, 9, 509.
- Huang, T.; Holden, J. A.; Heath, D. E.; O’Brien-Simpson, N. M.; O’Connor, A. J. Engineering Highly Effective Antimicrobial Selenium Nanoparticles Through Control of Particle Size. Nanoscale. 2019, 11(31), 14937–14951. DOI: 10.1039/C9NR04424H.
- Zhang, H.; Li, Z.; Dai, C.; Wang, P.; Fan, S.; Yu, B.; Qu, Y. Antibacterial Properties and Mechanism of Selenium Nanoparticles Synthesized by Providencia sp. DCX. Environ. Res. 2021, 194, 110630. DOI: 10.1016/j.envres.2020.110630.
- Chudobova, D.; Cihalova, K.; Dostalova, S.; Ruttkay-Nedecky, B.; Merlos Rodrigo, M. A.; Tmejova, K.; Kopel, P.; Nejdl, L.; Kudr, J.; Gumulec, J., et al. Comparison of the Effects of Silver Phosphate and Selenium Nanoparticles on Staphylococcus Aureus Growth Reveals Potential for Selenium Particles to Prevent Infection. FEMS Microbiol. Lett. 2014, 351(2), 195–201.
- Huang T.; Holden, J.; Heath, D.; O'Brien-Simpson, N.; O'Connor, A . Engineering Highly Effective Antimicrobial Selenium Nanoparticles Through Control of Particle Size. Nanoscale. 2019, 11(31), 14937–14951.
- Ghosal, A.; Nielsen, P. E. Potent Antibacterial Antisense Peptide–Peptide Nucleic Acid Conjugates Against Pseudomonas aeruginosa. Nucleic. Acid Ther. 2012, 22(5), 323–334. DOI: 10.1089/nat.2012.0370.
- Ebadi, M.; Zolfaghari, M. R.; Aghaei, S. S.; Zargar, M.; Shafiei, M.; Zahiri, H. S.; Noghabi, K. A. A Bio-Inspired Strategy for the Synthesis of Zinc Oxide Nanoparticles (ZnO NPs) Using the Cell Extract of Cyanobacterium Nostoc sp. EA03: From Biological Function to Toxicity Evaluation. Rsc. Adv. 2019, 9(41), 23508–23525. DOI: 10.1039/C9RA03962G.
- Varlamova, E. G.; Goltyaev, M. V.; Mal’tseva, V. N.; Turovsky, E. A.; Sarimov, R. M.; Simakin, A. V.; Gudkov, S. V. Mechanisms of the Cytotoxic Effect of Selenium Nanoparticles in Different Human Cancer Cell Lines. Int. J. Mol. Sci. 2021, 22(15), 7798. DOI: 10.3390/ijms22157798.