ABSTRACT
The field of epitranscriptomics is rapidly developing. Several modifications (e.g. methylations) have been identified for different RNA types. Current evidence shows that chemical RNA modifications can influence the whole molecule’s secondary structure, translatability, functionality, stability, and degradation, and some are dynamically and reversibly modulated. miRNAs, in particular, are not only post-transcriptional modulators of gene expression but are themselves submitted to regulatory mechanisms. Understanding how these modifications are regulated and the resulting pathological consequences when dysregulation occurs is essential for the development of new therapeutic targets. In humans and other mammals, dietary components have been shown to affect miRNA expression and may also induce chemical modifications in miRNAs. The identification of chemical modifications in miRNAs (endogenous and exogenous) that can impact host gene expression opens up an alternative way to select new specific therapeutic targets.
Hence, the aim of this review is to briefly address how RNA epitranscriptomic modifications can affect miRNA biogenesis and to summarize the existing evidence showing the connection between the (de)regulation of these processes and disease settings. In addition, we hypothesize on the potential effect certain chemical modifications could have on the potential cross-kingdom journey of dietary plant miRNAs.
Introduction
Emerging evidence implicates a wide range of post-transcriptional RNA modifications (epitranscriptome) that play crucial roles in fundamental biological processes, including the regulation of gene expression. Collectively, these modifications are known as epitranscriptomics.
Most RNAs families possess several co- or post-transcriptional chemical modifications at a variety of locations [Citation1,Citation2]. Comparable to DNA and histone modifications, it is expected that many of these RNA modifications may be associated with regulatory functions. Current evidence shows that chemical RNA modifications can influence the whole molecule’s secondary structure, translatability, functionality, stability and degradation, and some are dynamically and reversibly modulated [Citation2–8]. These modifications have been studied in the past decades mostly in messenger RNA (mRNA), though, more recently, many began to be described also in traditionally considered ‘functional’ (i.e. transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs)) and ‘non-functional’ non-coding RNAs (ncRNAs) (i.e. microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs)) [Citation9–16]. Understanding the biological consequences of such modifications is essential for the development of new therapeutic targets [Citation17,Citation18]. Although there are still many knowledge gaps, research in the field of epitranscriptomics is rapidly progressing and has been accelerated by the continuous advances in high throughput sequencing systems [Citation19].
MiRNAs are a class of single-stranded small (19–25 nucleotides) ncRNAs involved in the regulation of gene expression at a post-transcriptional level via mRNA silencing and translational repression, among other mechanisms [Citation20]. In humans and other mammals, dietary components have been shown to affect miRNA expression [Citation21,Citation22] and the likelihood that they may also induce chemical modifications in miRNAs, as was reported for mRNAs [Citation23], deserves further research. Indeed, the regulatory effect of miRNAs can also be transmitted between different species and kingdoms as a means of communication between two organisms [Citation24]. The identification of chemical modifications in miRNAs (endogenous and exogenous) – that can impact host gene expression – opens up an alternative way to select new specific therapeutic targets in different pathological settings.
Hence, the aim of this review is to briefly address how RNA epitranscriptomic modifications can affect miRNA biogenesis and to summarize the existing evidence showing the connection between the (de)regulation of these processes and disease settings. In addition, we hypothesize on the potential effect certain chemical modifications could have on the potential cross-kingdom journey of dietary plant miRNAs.
Epitranscriptomic modifications
The chemical structure of RNA can be modified in cells to serve as an epigenetic mechanism for gene expression control. Around 170 different chemical modifications of RNAs have been reported so far, a great number of them consist of a methyl (CH3) group being added to the nitrogenous base (e.g. N6-methyladenosine (m6A)), to the ribose sugar (e.g. 2′-O-methyladenosine) or to both (e.g. N6,2′-O-dimethyladenosine (m6Am)) [Citation1]. Many of these covalent RNA additions are highly prevalent and can be dynamically added and removed through writer and eraser complexes, respectively. In addition, these modifications are interpreted by modification-specific binding proteins known as readers, providing a new layer of epitranscriptome-mediated post-transcriptional regulation [Citation25]. The recent accumulation of knowledge has led to the development of RNA modification databases. For instance, MODOMICS database offers comprehensive evidence regarding the chemical structures of modified ribonucleosides, their biosynthetic pathways, the location of modified residues in RNA sequences, and RNA modifying enzymes [Citation1].
Adenosine modifications: N6-methyladenosine (m6A)
Adenosine methylation occurring at the sixth position of the nitrogenous base (N6-methyladenosine; m6A) is one of the most pervasive modifications found in eukaryotic RNA, including mRNA and ncRNA [Citation6,Citation26,Citation27]. m6A, which has been detected in animals, plants, single-cell organisms (archaea, bacteria and yeast) and viruses, has been linked to the regulation of multiple cellular processes, including RNA stability, translation, alternative splicing, secondary structure and nuclear export [Citation17,Citation28–36].
m6A writers, erasers and readers
The reversible addition and removal of methyl in N6 adenosine methylation is catalysed by methyltransferases (writers) and demethylases (erasers) proteins, respectively. In addition, methylated RNA binding proteins (readers) help regulate the downstream processes [Citation26]. The positions, patterns and motifs of m6A suggest that writers, readers and erasers might be conserved across kingdoms, though it has been suggested that individual members of m6A writer complex achieved functional divergence in plants [Citation37].
The association between methyltransferase-like (METTL) 3 and METTL14 with other regulator components, such as Wilm’s tumour-associated protein (WTAP), METTL16, zinc finger CCCH-type containing 13 (ZC3H13), and RNA binding motif protein (RBM) 15/15B, among others, results in a functional complex. This complex uses an S-adenosyl methionine (SAM) binding domain on METTL3 to methylate specific mRNAs at a RRACH (R = A/G; H = A/C/U) m6A consensus sequence, mainly in the 3′ untranslated regions (3ʹ UTRs) and near stop codons [Citation26,Citation38]. METTL3, the catalytic subunit, and METTL14, an allosteric activator, form a core heterodimer, whose localization in nuclear speckles is regulated by WTAP, which is also important for the methyl transferase activity of these enzymes [Citation39,Citation40]. WTAP-dependent binding between RBM15/15B and the methyltransferase complex is necessary for optimal methylation activity [Citation41]. ZC3H13 plays a critical role in anchoring WTAP and other components in the nucleus to facilitate m6A methylation [Citation38]. METTL16, known for its role as an RNA m6A methyltransferase, methylates mRNAs, including MAT2A, which encodes the SAM synthetase expressed in almost every cell [Citation34]. Although METTL16 is usually found in the nucleus, it has been suggested that it is also a cytoplasmic methyltransferase with different RNA binding features according to its cellular location [Citation42].
m6A was identified as a novel regulator of miRNA processing where METTL3 methylates primary inter- and intragenic miRNAs, and this modification was proposed to allow the microprocessor complex to recognize its specific substrates and initiate miRNA biogenesis [Citation43]. Moreover, METTL16 has been demonstrated to bind and methylate the U6 small nuclear RNA and the lncRNAs MALAT1 and XIST [Citation14,Citation34]. The fate of mRNAs containing m6A is predominantly determined by different categories of m6A-binding proteins, termed ‘readers’, which recognize the modified site and prompt downstream regulatory effects (e.g. tumorigenesis, viral replication, adipogenesis, haemopoiesis, immune regulation, etc.), by altering RNA metabolic processes [Citation25,Citation31–33,Citation44–46]. Such proteins include the YT521-B homology (YTH) domain family, heterogeneous nuclear ribonucleoproteins (hnRNPs), and IGF 2 mRNA-binding proteins (IGFBPs). The YTH domain represents a family that recognizes the m6A mark directly [Citation40], interacting with m6A present in RNA through a conserved aromatic (tryptophan) cage [Citation47,Citation48]. By recruiting different complexes to target m6A sites, the YTH domain-containing proteins, as well as other potential m6A-binding proteins, contribute to gene regulation post-transcriptionally in many aspects, such as splicing, translation, localization, and lifetime [Citation28,Citation29,Citation31,Citation33,Citation49]. The YTH domain family consists of YTH domain family protein 1–3 (YTHDF1-3, DF family) and YTH domain containing protein 1–2 (YTHDC1-2, DC family) [Citation49]. YTHDF1 protein can increment mRNA translation efficiency by way of interacting with the translation initiation factor eIF3 in a m7G-cap-independent way [Citation50]. The cytoplasmic m6A reader YTHDF2 is necessary for mRNA degradation and contributes to reducing the stability of targeted transcripts using the CCR4-NOT deadenylase complex [Citation28,Citation51]. YTHDF3 works in cooperation with these proteins to improve RNA binding specificity and affinity [Citation29,Citation32]. YTHDC1 and the YTHDF families are primarily localized to the nucleus and cytoplasm, respectively, while YTHDC2 is found in both the nucleus and cytoplasm [Citation31,Citation52,Citation53]. These proteins participate in the processes of gene splicing, exportation, degradation, and translation of m6A-containing mRNAs [Citation54–56]. Fragile-X mental retardation protein (FMRP) has been reported to be an indirect reader via binding with YTHDF proteins to regulate m6A-modified mRNA [Citation57].
The structural alteration of mRNAs induced by m6A methylation (m6A-switch) enhances the binding of other reader proteins such as heterogeneous nuclear ribonucleoproteins (hnRNPs), including hnRNPA2B1, hnRNPC and hnRNPG, and insulin-like growth factor 2 mRNA binding proteins (IGF2BPs), including IGF2BP1, IGF2BP2, and IGF2BP3, to m6A sites [Citation35,Citation36,Citation40]. In the m6A-switch mechanism, m6A modulates protein binding by inducing an RNA structural change that alters the accessibility of a protein binding site. In this sense, HNRNPC and HNRNPG binding to m6A-modified RNA depends on an m6A-induced base-pairing disruption that exposes U-rich and purine-rich binding sites, respectively [Citation35,Citation36,Citation58]. The former is involved in pre-mRNA processing, while HNRNPG plays a role in pre-mRNA alternative splicing. On the other hand, members -of the IGFBP family recognize m6A-containing transcripts via common RNA binding domains, such as the KH domain. Although the exact molecular mechanisms have not been fully revealed, IGF2BPs have been reported to recognize m6A-modified mRNAs and enhance their stability and translation. IGFBP proteins exert their functions by recruiting RNA stabilizers, such as HuR, to protect m6A-containing mRNA from degradation [Citation27,Citation30].
m6A modifications can be reversibly ‘erased’ in a dynamical process by RNA demethylases, such as fat mass and obesity-associated protein (FTO) and alkylation repair homolog protein 5 (ALKBH5) in mammals. The former is able to oxidize m6A resulting in intermediates, which are then hydrolysed to adenine, whereas ALKBH5 can eliminate m6A directly [Citation3,Citation27]. In plants, several ALKBH5 orthologs were detected, and it has been suggested that these proteins could have redundant functions in m6A demethylation, whereas no FTO orthologs were found [Citation59].
Other RNA modifications
Apart from m6A, various other RNA modifications have been identified so far. One of the most well-known chemical modifications of RNA affects capping at the 5′-end (5′ cap), including N7-methylguanosine (m7G) [Citation60,Citation61]. Other modifications, involving adenosine (e.g. N1-methyladenosine (m1A), N6,2′-O-dimethyladenosine (m6Am)), cytosine (e.g. 5-methylcytosine (m5C)), and uridine (e.g. uridylation, pseudouridylation), have been identified, as well as N-glycan alterations, among others. However, their biological roles are still not fully disclosed due to the complexity of RNA structure and functions [Citation27].
Adenosine modifications (other than m6A)
Other modifications of adenosine found in the human epitranscriptome include a methylation at the N1 position of adenosine, generating N1-methyladenosine (m1A), which is widespread in tRNA, rRNA and also in mRNA [Citation7,Citation62,Citation63]. Transcriptome-wide mapping of m1A exposed highly conserved features in mouse and human cells. In particular, these modifications were seen to reside close to both canonical and alternative transcription start sites (TSS), to be present in highly structured regions around the start codon, and to correlate positively with protein production [Citation62]. AlkB homolog 1, histone H2A dioxygenase (ALKBH1) was recently presented as an RNA demethylase that catalyzes the demethylation of m1A in tRNA [Citation64] and alkB homolog 3, alpha-ketoglutarate dependent dioxygenase (ALKBH3) has been suggested to be a potential demethylase of m1A in mRNA [Citation65].
In contrast to m6A, which is an internal modification, in higher eukaryotes, N6,2′-O-dimethyladenosine (m6Am) can be found in the first nucleotide after the 7-methylguanosine cap, near the TSS in certain mRNAs [Citation9,Citation65,Citation66]. Recently, m6Am was discovered to suppress cap-dependent translation, and PCIF1 was identified as mRNA m6Am methyltransferase [Citation67,Citation68].
Cytidine modifications
As mentioned above, apart from adenosine, the epitranscriptome exhibits several, increasingly characterized, modifications in other bases, such as cytidine. Most modifications are present at levels 10- to 100-fold less than m6A [Citation69]. Methylcytidine (m5C) modifications, identified in mRNAs and ncRNAs [Citation70], are present in modest levels (measured by LC–MS/MS) in mRNAs and have been mapped primarily to 5′ UTRs, whereas CDS regions showed the lowest density [Citation71,Citation72]. In humans, m5C in RNAs are catalysed by members 1–7 of the NOL1/NOP2/SUN domain (NSUN) family of proteins, together with the homologue of DNA methyltransferase (DNMT), DNMT2 [Citation10,Citation73]. NSUN6-methylation was recently associated with a role in regulating translation termination [Citation74].
Uridine modifications
3′ RNA uridylation, i.e. the addition of non-templated uridine(s) to the RNA end, by Terminal Uridylyl Transferases (TUTases), is now considered to be functionally important for RNA processing [Citation75]. Another important uridine-specific alteration is pseudouridylation, which consists in the creation of the 5-ribosyl isomer of uridine, pseudouridine (Ψ) [Citation76]. Ψ modifications relative to U in the 3′-UTR were distinctly lower compared to the 5′-UTR and CDS region in mammals, including humans [Citation77–79]. Pseudouridylation is catalysed by pseudouridine synthases (PUS), which have been linked to human diseases and important biological processes [Citation7].
RNA capping modifications
N7-methylguanosine (m7G) is a very-well described cap modification crucial for mRNA stability, translation and functional diversity [Citation61,Citation80,Citation81]. Despite its relative abundance, it is challenging to study since it does not interfere with reverse transcription (as it is neutral to Watson–Crick base pairing) and cannot be detected by standard sequencing-based technologies [Citation15].
N-glycan modifications
Glycans, which have been widely described in proteins and lipids, regulate a myriad of essential functions and are present across different kingdoms of life [Citation82]. Very few RNAs have been previously described to have sugar modifications in the side chain, including tRNA [Citation83]. A recent study showed that small noncoding RNAs contain sialylated glycans (‘glycoRNAs’) and that the majority of cellular glycoRNAs are located on the cell surface [Citation84]. These glycoRNAs seem to act as ligands of the sialic acid binding-immunoglobulin lectin-type (Siglec) receptors, which have been functionally associated with diseases, such as autoimmunity and cancer [Citation84]. Although more research is needed on this topic, the existence of glycoRNAs expands the role of RNA modification in health and disease.
Epitranscriptomic modifications in plants
RNA modifications such as m6A, m5C and Ψ have been detected in transcriptome-wide studies in plants. m6A sites are widespread in plants and exhibit the dominant m6A consensus motif RRACH (R = A/G; H = A/C/U), as well as other lineage-specific m6A motifs, such as URUAY [Citation46,Citation85,Citation86]. Throughout the transcriptome, m6A are more abundant near the stop codon and 3′ UTR in different developmental stages in Arabidopsis and other plants. Furthermore, several studies have shown m5C throughout the Arabidopsis transcriptome [Citation87,Citation88]. Other mRNA modifications, such as m1A, hm5C, Nm, I and ac4C have vital functions in mammals, but need to be further depicted in plants [Citation59].
Gene-expression levels of writers, erasers and readers vary across different plant tissues and developmental stages [Citation87,Citation89,Citation90], suggesting the dynamic regulation of mRNA modifications in plants. Furthermore, the level of mRNA modifications in different plant species has revealed the dynamics of mRNA modifications during plant growth and development [Citation91]. Even though a number of m6A writers, erasers and readers have been identified in Arabidopsis, the exact molecular mechanism describing the dynamic m6A generation and deletion in plants in response to environmental changes is still lacking.
The m6A methyltransferase complex seems to be conserved between mammals and plants [Citation92]. The core component with m6A catalytic activity is METTL3, which belongs to the MT-A70 family. In plants, the MT-A70 family proteins can be grouped in MTA, MTB, and MTC, which are present in most plant species [Citation93]. In Arabidopsis, the m6A writer complex contains the METTL3 ortholog, MTA [Citation89], and the METTL14 ortholog, MTB [Citation94], among other components. Many functional sites are conserved between plants and humans, suggesting that plant MTA–MTB and human METTL3–METTL14 may have a similar mechanism of methylation. As mentioned earlier, the methylation of A to m6A can be reversed by m6A demethylases (erasers). In plants, even though no FTO orthologs are found, genomes encode multiple copies of ALKBH5 orthologs, including six orthologs (ALKBH9A/B/C and ALKBH10A/B/C) in Arabidopsis, suggesting these may have redundant functions in m6A demethylation [Citation95]. Moreover, m6A mediates its biological function by recruiting reader proteins. Plant YTH proteins can be divided into two groups: group 1 includes YTHDF and the majority of YTH domain proteins, and group 2 contains YTHDC1, YTHDC2 and the remaining plant YTH proteins, which form two subgroups [Citation45]. The residues involved in the cage and RNA binding are highly conserved between yeast and Arabidopsis YTH proteins, suggesting that the other YTH proteins in Arabidopsis could also be m6A readers.
In mammals, m6A is involved in almost all aspects of RNA metabolism, including transcript stability, translation initiation, mRNA export, polyadenylation, nuclear retention, and pre-mRNA splicing [Citation2,Citation3,Citation5,Citation6,Citation54]. In plants, several studies have demonstrated that m6A and m5C affect RNA stability [Citation46,Citation85,Citation87,Citation90,Citation95]. In contrast, a number of transcripts exhibited increased abundance after loss of m6A in plants [Citation85,Citation86,Citation90,Citation95], raising the question of how m6A regulates gene expression in different plant species and under specific circumstances.
In humans, m5C RNA modifications are catalysed by NSUN2 [Citation44,Citation70], whereas in plants, its homolog tRNA-specific methyltransferase 4B (TRM4B), belonging to the RNA (C5-cytosine) methyltransferase (RCMT) family, functions as an m5C mRNA methyltransferase in Arabidopsis [Citation87,Citation88]. In plants, m5C seems to have an ancient origin and widespread distribution, since members of the RCMT family are present in a great number of plant species, ranging from green algae to flowering plants.
miRNAs biogenesis, target recognition and biological functions
As mentioned above, miRNAs are a class of single-stranded non-coding RNAs, highly conserved evolutionarily, which have been associated with key regulatory roles in a substantial number of cellular processes in eukaryotic cells [Citation96].
The biogenesis of miRNA is classified into canonical and non-canonical pathways. Briefly, the canonical pathway is characterized by a preliminary cleavage of primary miRNA (pri-miRNA) to precursor miRNA (pre-miRNA) by the microprocessor complex consisting of Drosha and DGCR8 enzymes. Following its transfer to the cytoplasm by exportin 5, pre-miRNA is additionally cleaved by Dicer to generate double-strand RNAs, which are subsequently associated with Argonaute (AGO) proteins forming the RNA-induced silencing complex (RISC) [Citation97]. Non-canonical pathways circumvent the steps of the canonical pathway and can be differentiated into Drosha/DGCR8-independent and Dicer-independent pathways [Citation98].
It is acknowledged that miRNAs can directly mediate post-transcriptional gene silencing in the cytoplasm through a seed sequence, a run of six nucleotides spanning nucleotide 2–7 on the 5′ end of miRNAs, and the complementary sequences in the 3′-untranslated region (UTR) of target mRNA [Citation99]. Nevertheless, the interaction between miRNAs and other regions, such as 5′ UTR, coding sequence, and gene promoters, have also been reported [Citation100]. In addition, it seems that miRNAs can be shuttled between different subcellular compartments to control the rate of translation and transcription [Citation101]. Recent studies have shed light on the dynamic nature of miRNA actions and further revealed the complexity of miRNA-mediated gene regulation [Citation102]. miRNA actions are vital for animal development and viability, and the deregulation of their activity has often been associated with the development and progression of human diseases [Citation103].
Crosstalk between RNA modifications and miRNA in disease settings
Evidence of the crosstalk between RNA modifications and miRNA biogenesis in pathophysiological contexts is rapidly growing. Examples of these associations are summarized in .
Table 1. Summary of studies reporting a crosstalk between RNA modifications and miRNAs in disease settings
Adenosine modifications, miRNAs and disease
Significant overlap between m6A and miRNA-binding sites has been reported in 3ʹ UTRs of mRNAs, and this co-localization generally shows an inverse allocation pattern, where m6A peaks abound near the stop codon, usually preceding miRNA-binding sites, which prosper near the 3ʹ end of 3ʹ UTRs [Citation104]. The inverse distribution pattern seems to indicate that the interaction between m6A and downstream-bound miRNA requires a particular partition. Furthermore, raised expression of miRNAs was frequently associated with larger amounts of m6A-modified target transcripts, suggesting that miRNAs may regulate m6A modifications on their target transcripts [Citation104]. In addition, it has been reported that m6A marks the initiation of pri-miRNAs processing [Citation5]. HNRNPA2B1 was categorized as m6A-reader protein in the nucleus that interacts with the Microprocessor protein DGCR8, enhances binding of DGCR8 to pri-miRNA transcripts, and positively regulates pri-miRNA processing in a similar manner as METTL3 [Citation5,Citation43]. Moreover, HNRNPA2B1 modulates the alternative splicing of mRNA transcripts [Citation43]. Recent studies have provided evidence on the link between m6A modification, miRNAs processing and tumour progression scenarios. As previously described, METTL3, in an m6A-dependent manner, interacts with DGCR8 to support the maturation of pri-miR221/222, which then targets PTEN, leading to accelerated cell proliferation in bladder cancer [Citation105]. In colorectal cancer, METTL3 can methylate pri-miR-1246, promoting the maturation of pri-miR-1246, which targets the anti-oncogene SPRED2 [Citation106]. Moreover, METTL3 was shown to prompt the splicing of pre-miR-143-3p into its mature form, which is involved in the brain metastasis of lung cancer cells via down regulation of VASH1 [Citation107]. In a recent investigation, cigarette smoke-induced overexpression of METTL3 resulted in increased m6A modification, with the involvement of NF-κB associated protein (NKAP). In turn, this generated excessive maturation of pri-miR-25, which impairs PH domain leucine-rich repeat protein phosphatase 2 (PHLPP2), contributing to the activation of AKT-p70S6K signalling, a pathway linked to the development and progression of pancreatic cancer [Citation108]. In another study, METTL14 has been shown to interact with DGCR8 enhancing the maturation of pri-miR-126, which then counteracts the suppressing impact of this m6A writer on the metastasis in hepatocellular carcinoma (HCC) [Citation109]. Evidence, concerning other pathological scenarios associated with irregular m6A-dependent miRNA processing, is beginning to accumulate. For example, it has been reported that METTL3 interacts with DGCR8 and stimulates the maturation of miR-873-5p, which could regulate Keap1-Nrf2 pathway against oxidative stress and apoptosis in colistin-evoked nephrotoxicity [Citation110]. In addition, METTL3-catalyzed m6A resulted in a negative regulation of pre-miR-320 and miR-320, which targets RUNX2, a key transcription factor for osteoblast differentiation and bone formation [Citation111]. In gastrointestinal cancer, Konno and colleagues reported a rise in the levels of METTL3 and METTL14 together with a higher number of RNA methyl marks, including m6A, in miR-200 c, miR-17-5p, let-7a-5p, among other miRNAs [Citation112]. Lastly, METTL3‐dependent m6A was shown to regulate miR‐7212‐5p maturation via DGCR8, a microRNA that targets FGFR3 to inhibit osteoblast differentiation in mouse osteoblast precursor cells [Citation113].
Accumulating evidence supports the concept that m6A influences miRNAs processing, yet it does not appear to play a unidirectional regulation as the role of miRNAs in regulating m6A formation of mRNAs has been revealed [Citation13]. Whereas the involvement of AGO proteins in regulating m6A formation was discarded, this was not the case for Dicer, whose induction raised the abundance of m6A without altering the protein levels of METTL3, FTO and ALKBH5. In addition, Dicer seems to influence the nuclear speckle localization of METTL3 [Citation13]. Overexpression and knockdown of specific miRNAs increased and decreased, respectively, m6A abundance in mouse neural crest stem cells (NSCs) and in HeLa cells. Finally, miRNAs were shown to regulate the m6A methyltransferase activity of METTL3 by modulating its binding to mRNAs [Citation13]. The above is supported by a study performed in HCC cells, where miR-145 was reported to regulate m6A level through the inhibition of YTHDF2 expression [Citation114]. In another study, focused on hepatoblastoma progression, miR‐186 was proposed to control METTL3 expression via Wnt/β‐catenin signalling [Citation115]. Moreover, the existence of a feedback loop was proposed, wherein HBXIP suppresses let-7 g inducing METTL3, which in turn increases HBXIP expression, leading to the acceleration of proliferation in breast cancer [Citation116]. Regarding the RISC complex structural changes influenced by methylated miRNAs, in vitro and in silico analyses suggest that m6A modifications in miR-17-5p and let-7a-5p, located away from the RNA-binding site (at positions 13 and 19, respectively) affect target-RNA recognition efficiency [Citation112]. These results demonstrate that m6A modifications may reduce the ability of miRNAs to suppress target mRNA translation. Nevertheless, m6A-modified miR-200 c-3p did not repress target gene expression, contrary to non-methylated and m5C-modified miR-200 c-3p in HCT116, a low-level endogenous miRNA expressing cell line [Citation112].
To the best of our knowledge, solid evidence on the impact of m1A on the processing of a specific miRNA has not yet been reported in a pathological context. Nevertheless, m1A modification sites have been identified in lncRNA, such as MALAT1 (m1A8398), which was found to be upregulated in various forms of cancer [Citation117], and in a group of nuclear-encoded lncRNAs [Citation118].
Cytidine modifications, miRNAs and disease
In 2019, Konno et al. showed that miRNAs could be cytosine-methylated and reported that adenosine and cytosine methylated miRNA-17-5p could be used as a biomarker of early-stage pancreatic cancer [Citation112]. In gastric cancer cell lines, small RNA methyl marks, including 5mC, 3-methylcytosine, m1A and m6A, were found in 1–8% of total adenines and cytosines. Interestingly, the fraction of methylated miRNAs increased upon stimulation with epidermal growth factor, suggesting a regulatory mechanism for RNA modification [Citation112]. Cheray et al. (2020) confirmed the existence of cytosine methylation in mature miRNAs and show it is associated with poor prognosis in glioblastoma multiforme [Citation119]. In addition, this study provided evidence that miRNAs can be cytosine-methylated by DNMT3A/AGO4-including complexes and demonstrated that 5mC in miRNAs suppresses their gene expression repressive capacity [Citation119]. Very recently, Carissimi and colleagues confirmed the presence of m5C in human miRNAs and reported the existence of (hydroxy)-methyl-5 cytosine (hm5C) modifications [Citation16]. In human cell lines and PBMCs, these authors found that several miRNAs (i.e. miR-125a-5p, miR-191-5p, miR-25-3p and many others) harbour hm5C and that this is not restricted to CG sequence context. These findings suggest that, in addition to DNMT3A, other RNA methyltransferases might be involved, although the functional consequences of these modifications are unknown [Citation16]. Another RNA cytosine modification, ac4C, is regulated by N-acetyltransferase 10 (NAT10). Ac4C is enriched within the 5ʹ regions of the coding sequence and is associated with substrate mRNA stability [Citation8].
Uridine modifications, miRNAs and disease
It is known that uridylation plays a role in the regulation of canonical microRNA biogenesis in the tumour suppressor let-7 microRNA family. In addition, uridulation involves two different non-canonical microRNA biogenesis pathways contrasting on their dependence on enzymes Drosha and Dicer. Furthermore, uridylation of mature miRNAs can generate isomiRs, sequence variant microRNAs, with modified action [Citation75]. TUT1 has been suggested to positively influence, through 3′ nucleotide additions, the levels of miR-24 and miR-29a, which, in turn, negatively modulate lipogenesis regulators PPAR-gamma and SREBP-1 c. As lipogenesis is considered to be a cancer hallmark, TUT1 might act as a tumour suppressor [Citation120]. miRNAs play important roles in disease progression, mainly through the regulation of their target genes; however, miRNAs can themselves be post-transcriptionally regulated by other genes, which contributes to the overall complexity of the regulation process. A clear example occurs with the regulation of the Let-7 miRNA family biogenesis through pre-miRNA uridylation, where TUTases 4 and 7 interacts with Lin28, which, in turn, is regulated via Let-7 miRNA, generating a negative feedback loop [Citation12,Citation121]. In head and neck cancer [Citation122], and also in breast cancer [Citation123], a suppression mediated by LIN28 was observed in the processing of pre-let-7. Notably, in the former study, this was accompanied by the up-regulation of genes in the IGF pathway in Lin28b-expressing cells [Citation122], namely IGF2BP2, while in the latter study the influence of Lin28 on let-7 maturation was associated to the Wnt–b-catenin pathway [Citation123]. DIS3-like exonuclease 2 (DIS3L2) was proposed to be the reader for uridylated pre-let-7 in vivo, forming a LIN28-TUT4/7-DIS3L2 pathway [Citation124]. However, it was recently reported that DIS3L2 loss had no effect on mature let-7 levels [Citation125].
Most studies of PUSs and Ψ have focused on abundant ncRNAs, and evidence regarding whether PUSs are able to influence metabolic pathways of other types of RNA is scarce. Recently, Song et al. (2020) reported that the depletion of PUS10 caused a noticeable decrease in the expression of many mature miRNAs together with the build-up of unprocessed pri-miRNAs in several human cells [Citation126]. This study also showed that PUS10 directly binds to pri-miRNAs and interacts with the microprocessor to promote miRNA biogenesis; however, authors concluded that the catalytic activity of PUS10 does not play a relevant role in this process [Citation126].
RNA capping modifications, miRNAs and disease
Recently, two complementary high-throughput sequencing strategies were used to demonstrate that a subset of miRNAs harbours internal m7G modification, including the let-7 family. In particular, it was shown that m7G methylation by METTL1, a methyltransferase that regulates cell migration and promotes the processing of let-7e-5p miRNA precursor [Citation15]. Apart from m7G, other somewhat less characterized RNA caps exist in eukaryotes (e.g. yeasts, plants and humans) [Citation4,Citation127–129]. The recognition of the 5′mono-phosphate of pre-miRNAs by Dicer is important for the subsequent processing of miRNAs. In this sense, a study by Xhemalce et al. (2012) showed that the BCDIN3 domain containing RNA methyltransferase (BCDIN3D) was shown to O-methylate the 5′ mono-phosphate end of pre-miR-145 and pre-miR-23b, inhibiting their processing by Dicer [Citation127].
3′ end modifications affect the regulation of miRNA stability and target recognition
As mentioned above, miRNA are important regulators of many vital cellular processes; hence, turnover and degradation of these regulators needs to be tightly and dynamically controlled. This can be achieved through the manipulation of miRNA stability, which involves 3′ end modifications (i.e. 3′ methylation and 3′-to-5′ truncation), AGO association, and miRNA-target RNA interaction. Extended information on these mechanisms can be found elsewhere [Citation130]. Regarding 3′ end modifications, the HEN1-catalysed 2′-O-methylation modification at the 3′ end (3′-terminal 2′Ome) of various small RNAs (including miRNAs) in plants and piRNAs in animals is crucial for their stability (). Loss of function mutations involving hen1 cause miRNA deterioration and 3′ end heterogeneity due to combined actions of 3′-to-5′ truncation and 3′ tailing, mainly by uridylation [Citation11,Citation131,Citation132]. The addition of non-uridine nucleotides to miRNAs was also shown to occur at very low frequencies through other non-identified terminal nucleotidyl transferases. However, their biological relevance (if any) remains to be ascertained [Citation132].
Figure 1. Epitranscriptomic modifications in miRNAs. Schematic representation of miRNA biogenesis in mammals and plants. HEN-1 is a methyltransferase that adds a methyl group to the ribose of the last nucleotide (represented as R) of small RNAs (sRNAs). This methylation protects the 3ʹ-end of sRNAs from uridylation activity and subsequent degradation. Not all miRNA biogenesis mechanisms are depicted. Frequently reported miRNA modifications, along with representative examples, are shown on the right bottom corner
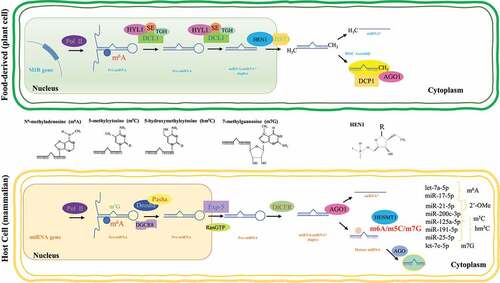
In Arabidopsis, HEN1 Suppressor 1 (HESO1) is an important TUT that catalyzes the addition of uracils to the 3′ end of unmethylated miRNAs [Citation131]. UTP:RNA uridylyltransferase 1 (URT1) is another TUT reported to be a functional paralog of HESO1 [Citation132,Citation133]. URT1 and HESO1 act cooperatively on AGO1-bound miRNAs and the tailed miRNAs stay bound by AGO1. Moreover, miRNA tailing affects the activity of miRNAs, in addition to causing miRNA degradation [Citation133]. In addition, small RNA Degrading Nucleases (SDNs), which are 3′ to 5′ end exoribonucleases, contribute to miRNA degradation by 3′ truncation of AGO1-bound and methylated miRNAs to result in AGO1-bound, 3′ truncated-and-unmethylated miRNAs. These are then uridylated by HESO1 and/or URT1, and further degradation is catalysed by other enzymes [Citation134,Citation135]. Small RNA degradation associated with uridylation seems to be a conserved mechanism in a variety of species, as is the case of the green algae Chlamydomonas reinhardtii, where MUT68, a terminal nucleotidyl transferase homologous to HESO1, is involved in the untemplated uridylation of the 3′ end of miRNAs and siRNAs [Citation136].
Recently, HEMT1, an HEN1 homolog, was identified as the methyltransferase responsible for 3′-terminal 2′Ome of mammalian miRNAs [Citation137]. Moreover, HENMT1 was significantly increased in lung cancer tissues compared to non-cancerous lung tissues, and differential 3′-terminal 2′Ome was shown for miR-21-5p in non-small cell lung cancer (NSCLC) and miR-26-5p in non-cancerous lung tissues. Furthermore, in vitro cleavage assays indicated that 3′-terminal 2′Ome protected miRNA 3′→5′ degradation by PNPT1. In addition, methylated miR-21-5p exhibited a higher affinity to AGO2 than the unmethylated form. miRNA target recognition should not be affected by 3ʹ uridylation since alterations at the 3ʹ end of miRNAs do not alter its seed sequence. However, it was recently shown that TUT4/7-mediated uridylation of miR-27a can modulate its pairing, enabling it to repress a distinct set of targets, despite the absence of a seed pairing match [Citation138].
Despite recent advances, further research is needed to understand how the (de)regulation of 3′-terminal methylations in mammalian miRNAs is associated with disease settings.
Epitranscriptomic modifications potential contribution to cross-kingdom regulation
In recent decades, research has found that miRNA signals can be transmitted between different species and kingdoms as a means of communication between two, often unrelated, interacting organisms, such as a host and a pathogen, pest, parasite, or symbiont [Citation24,Citation139]. The identification of food-derived miRNAs from animal [Citation140] and plant kingdoms [Citation141] brought attention to the possibility that dietary miRNAs could influence host gene expression [Citation142,Citation143]. Nevertheless, even though several studies have been reported since the first paper by Zhang et al. (2012), kingdom crossing of food-derived miRNAs is far from being a consensual matter [Citation144,Citation145].
Before any biologically meaningful effects may eventually occur, dietary miRNAs must surpass many obstacles before (food processing-related) and after (bioavailability, reaching and entering target cells, etc.) entering host organisms [Citation143]. First, miRNAs stability must be maintained throughout the harsh food processing chain (e.g. ripening, storage, cooking and/or other technological practices). Preliminary studies suggest that the stability of miRNAs depends on their specific sequence (i.e. GC content) [Citation146] and secondary structure [Citation147]. Specific dietary miRNAs have been reported to remain highly stable during different handling, storage and/or cooking conditions [Citation141,Citation148] or to resist degradation even under harsh conditions, such as acidic pH and substantial heat exposure (e.g. boiling) [Citation149,Citation150]. For example, MIR2911, a plant 26S ribosomal RNA-derived small RNA, exhibited high stability against RNases and strong stability in circulation[Citation150]. However, many miRNAs do not survive food processing intact and certain conditions, such as high temperature and pressure, starch dextrinization or protein denaturation, reduce their contents [Citation151,Citation152]. For example, in animal-derived products, such as milk and its derivatives, processing (raw milk pasteurization and homogenization) leads to significant miRNA loss [Citation153]. In plant-based foods, such as olive oil or beer, miRNAs are barely detected, most likely as a consequence of extensive degradation due to cellular compartment rupture and long-term contact between miRNAs and RNases [Citation154]. Regarding the harsh biological factors miRNAs face after entering the host organism, sequence [Citation146] and secondary structure [Citation147] also influence their stability. The unfavourable environment of the gastrointestinal tract, including RNases and lytic enzymes of the oral cavity, extreme stomach acidity, flow of gastroenteric fluid and peristaltic activity, nucleases and degradative enzymes, mucus composition and gut microbiota, are conditions that dietary miRNAs must surpass before they can be taken up, biodistributed to target tissues and, eventually, exert biological effects [Citation142,Citation143]. In this sense, some studies failed to detect the transfer of plant or exosomal miRNAs Citation155,Citation156 .
Although it is yet to be demonstrated in vivo, dietary miRNAs may cross the intestinal epithelium using the available routes of absorption (i.e. paracellular or transcellular). Some stable plant miRNAs can be absorbed via clathrin- and caveolin-mediated endocytosis [Citation147]. In vitro, miRNA uptake was observed to be sequence-dependent and facilitated by NACh and TLR9 cell membrane receptors [Citation147]. The absorption of single-stranded dietary miR168a seems to be facilitated compared to other types of miRNA structure [Citation141], while the uptake of single-stranded mature miRNA was significantly lower than that of double-stranded miRNA mimics at pH 7.4 in cultured cells [Citation157]. However, the absorption of dietary miRNAs by gastric pit cells in the stomach has been reported to be pH-dependent, as a near 30-fold rise was seen in single-stranded miR2911 absorption in pH conditions of 3.5 compared to 7.4 [Citation157]. Although it is not clear whether the epitranscriptomic modifications of miRNAs influence their dietary absorption, the stability increasing effects (described above) may increase survival chances. Increased absorption of dietary small RNAs due to alterations in intestinal permeability have been reported [Citation146]. miRNA packaging in extracellular vesicles (EVs), followed by their release in the circulatory system, enhances nuclease resistance and facilitates their uptake compared to the unpackaged form [Citation141]. Why and how dietary miRNAs are packaged into mammalian EVs remains to be determined. Once taken up by the target cells/tissues, the biological functions potentially exerted by dietary miRNAs will depend on factors, such as subcellular localization [Citation158], number of target transcripts [Citation159], and/or concentration reached within the cell [Citation160].
There have been very few studies where the mechanisms of molecular interaction between human AGO proteins and plant miRNAs within the RISC complex, where miRNA functions have been explored. It has been shown that 3′-terminal 2′Ome (but not at the 5ʹ termini) of miRNAs largely protects them from nuclease degradation, suggesting that miRNAs are likely to be destabilized at their 3΄ ends upon target binding [Citation161]. It is also well known that 3ʹ-end methylation is a common step in miRNA and siRNA metabolism and expectedly protects the 3ʹ ends of small RNAs from the uridylation activity and trimming in plants [Citation11]. Moreover, HENMT1-mediated 3′-terminal 2′Ome of miRNAs in humans seems to increase their higher affinity with the AGO2 complex and provide some protection against degradation [Citation137]. In this sense, it cannot be discarded that certain chemical modifications of dietary plant miRNAs provide them with enhanced protection against degradation by host organisms, and that these miRs could, eventually, reach target cells in sufficient numbers for biological effects to occur.
The hypothesis of cross-kingdom regulation of gene expression by dietary miRNAs represents a novel approach for future dietary therapy and deserves further research [Citation142,Citation145]. However, the theory of cross-kingdom regulation leading to relevant biological effects has been refuted by many researchers, and there are still many loopholes to fulfill and risks to consider before the use of dietary or plant miRNAs as effective therapeutic tools can be a reality.
Future Perspectives
Aberrant expression of ncRNAs, particularly miRNAs, is found in pathological contexts. In recent times, target selection in the development of miRNA-based therapeutics is mainly based on quantitative criteria (expression studies) and less on qualitative criteria (e.g. presence/absence of specific chemical modifications) [Citation18,Citation162,Citation163]. RNA modifications offer a novel layer of complexity to the intricate mechanisms of disease development and therapy. Synchronously to their functioning as post-transcriptional mediators, miRNA biogenesis and target recognition can be regulated through chemical modifications. New specific therapeutic targets can be selected by studying chemically modified miRNAs. For example, uridylation of miR-26a prevents the miRNA from repressing its mRNA target, without affecting its abundance [Citation164]. The growing knowledge in the epitranscriptomics field needs to continue in order to achieve the ultimate goal of providing the bases for the implementation of dependable, minimally invasive, diagnostic and therapeutic tools in the clinical setting.
On the other hand, it is important to determine if miRNA modifications can have an impact on cross-kingdom regulation (e.g. by enhancing miRNA stability) and contribute to potentiate the development of therapeutic miRNAs within edible plants. In this context, potent practical applications based on RNA oligonucleotides heavily rely on the inclusion of chemical modifications to exert enhanced biological effects [Citation165]. Moreover, the development of genetically modified crops containing miRNAs is guaranteed [Citation146,Citation166,Citation167,Citation168].
Author contribution
AD, JT-C and MCLH designed the manuscript and contents. JT-C, AD and MCLH wrote and revised the first version of the manuscript. HB, YB, AC and MMG revised and edited the second version of the manuscript. All authors revised and approved the final version.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018 Jan 4;46(D1):D303–d307.
- Boo SH, Kim YK. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med. 2020 Mar;52(3):400–408.
- Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014 May;15(5):313–326.
- Abdelhamid RF, Plessy C, Yamauchi Y, et al. Multiplicity of 5ʹ cap structures present on short RNAs. PloS One. 2014;9(7):e102895.
- Alarcón CR, Lee H, Goodarzi H, et al. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015 Mar 26;519(7544):482–485.
- Wu R, Jiang D, Wang Y, et al. N (6)-Methyladenosine (m(6)A) Methylation in mRNA with A dynamic and reversible epigenetic modification. Mol Biotechnol. 2016 Jul;58(7):450–459.
- Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017 Jun 15;169(7):1187–1200.
- Arango D, Sturgill D, Alhusaini N, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018 Dec 13;175(7):1872–1886.e24.
- Wei C, Gershowitz A, Moss B. N6, O2ʹ-dimethyladenosine a novel methylated ribonucleoside next to the 5ʹ terminal of animal cell and virus mRNAs. Nature. 1975 Sep 18;257(5523):251–253.
- Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999 Aug 1;27(15):3138–3145.
- Li J, Yang Z, Yu B, et al. Methylation protects miRNAs and siRNAs from a 3ʹ-end uridylation activity in Arabidopsis. Curr Biol. 2005 Aug 23;15(16):1501–1507.
- Heo I, Joo C, Kim YK, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009 Aug 21;138(4):696–708.
- Chen T, Hao YJ, Zhang Y, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015 Mar 5;16(3):289–301.
- Brown JA, Kinzig CG, DeGregorio SJ, et al. Methyltransferase-like protein 16 binds the 3ʹ-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci U S A. 2016 Dec 6 113(49):14013–14018.
- Pandolfini L, Barbieri I, Bannister AJ, et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell. 2019 Jun;20;74(6):1278–1290.e9.
- Carissimi C, Laudadio I, Lorefice E, et al. Bisulphite miRNA-seq reveals widespread CpG and non-CpG 5-(hydroxy)methyl-Cytosine in human microRNAs. RNA Biol. 2021 Jun;7:1–10.
- Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014 Dec;24(12):1403–1419.
- Torsin LI, Petrescu GED, Sabo AA, et al. Editing and chemical modifications on Non-Coding RNAs in Cancer: a new tale with clinical significance. Int J Mol Sci. 2021 Jan 8;22:2.
- Tk D, Xhemalçe B. Deciphering RNA modifications at base resolution: from chemistry to biology. Brief Funct Genomics. 2021 Mar 27;20(2):77–85.
- Dávalos A, Pinilla L, M-c LDLH, et al. Dietary microRNAs and cancer: a new therapeutic approach? Semin Cancer Biol. 2021 2021/08/01/;73:19–29.
- Mantilla-Escalante DC, López de Las Hazas MC, Crespo MC, et al. Mediterranean diet enriched in extra-virgin olive oil or nuts modulates circulating exosomal non-coding RNAs. Eur J Nutr. 2021 May 23;60(8):4279–4293.
- López de Las Hazas MC, Gil-Zamorano J, Cofán M, et al. One-year dietary supplementation with walnuts modifies exosomal miRNA in elderly subjects. Eur J Nutr. 2021 Jun;60(4):1999–2011.
- Lu N, Li X, Yu J, et al. Curcumin Attenuates Lipopolysaccharide-Induced hepatic lipid metabolism disorder by modification of m(6) A RNA methylation in piglets. Lipids. 2018 Jan;53(1):53–63.
- Weiberg A, Bellinger M, Jin H. Conversations between kingdoms: small RNAs. Curr Opin Biotechnol. 2015 Apr;32:207–215.
- Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017 Oct 6;33:319–342.
- Karthiya R, Khandelia P. m6A RNA methylation: ramifications for gene expression and human health. Mol Biotechnol. 2020 Oct;62(10):467–484.
- Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020 Feb 28;19(1):44.
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014 Jan 2;505(7481):117–120.
- Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017 Mar;27(3):315–328.
- Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018 Mar;20(3):285–295.
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015 Jun 4;161(6):1388–1399.
- Li A, Ys C, Xl P, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017 Mar;27(3):444–447.
- Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016 Feb 18;61(4):507–519.
- Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m(6)A Methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017 May 18;169(5):824–835.e14.
- Liu N, Dai Q, Zheng G, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015 Feb 26;518(7540):560–564.
- Liu N, Zhou KI, Parisien M, et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017 Jun 2;45(10):6051–6063.
- Kumar S, Mohapatra T. Deciphering Epitranscriptome: modification of mRNA bases provides a new perspective for post-transcriptional regulation of gene expression. Front Cell Dev Biol. 2021;9:628415.
- Wen J, Lv R, Ma H, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018 Mar 15;69(6):1028–1038.e6.
- Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016 Jul 21;63(2):306–317.
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019 Oct;20(10):608–624.
- Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016 Sep 15;537(7620):369–373.
- Nance DJ, Satterwhite ER, Bhaskar B, et al. Characterization of METTL16 as a cytoplasmic RNA binding protein. PloS One. 2020;15(1):e0227647.
- Alarcón CR, Goodarzi H, Lee H, et al. HNRNPA2B1 Is a Mediator of m(6) A-dependent nuclear RNA processing events. Cell. 2015 Sep 10;162(6):1299–1308.
- Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017 May;27(5):606–625.
- Scutenaire J, Deragon JM, Jean V, et al. The YTH domain protein ECT2 Is an m(6)A reader required for normal trichome branching in arabidopsis. Plant Cell. 2018 May;30(5):986–1005.
- Wei LH, Song P, Wang Y, et al. The m(6)A Reader ECT2 controls trichome morphology by affecting mRNA stability in arabidopsis. Plant Cell. 2018 May;30(5):968–985.
- Xu C, Liu K, Ahmed H, et al. Structural basis for the discriminative recognition of N6-Methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. 2015 Oct 9;290(41):24902–24913.
- Theler D, Dominguez C, Blatter M, et al. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014 Dec 16;42(22):13911–13919.
- Liao S, Sun H, Xu C, et al. A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinformatics. 2018 Apr;16(2):99–107.
- Meyer KD, Patil DP, Zhou J, et al. 5ʹ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015 Nov5;163(4):999–1010.
- Du H, Zhao Y, He J, et al. YTHDF2 destabilizes m(6) A-containingRNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016 Aug 25;7(1):12626.
- Hartmann AM, Nayler O, Schwaiger FW, et al. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol Biol Cell. 1999 Nov;10(11):3909–3926.
- Wojtas MN, Pandey RR, Mendel M, et al. Regulation of m(6)A Transcripts by the 3ʹ→5ʹ RNA Helicase YTHDC2 Is essential for a successful meiotic program in the mammalian germline. Mol Cell. 2017 Oct 19;68(2):374–387.e12.
- Roundtree IA, He C. Nuclear m(6)A Reader YTHDC1 regulates mRNA splicing. Trends Genet. 2016 Jun;32(6):320–321.
- Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6) A-BindingProteins. Trends Cell Biol. 2018 Feb;28(2):113–127.
- Mao Y, Dong L, Liu XM, et al. m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun. 2019 Nov25;10(1):5332.
- Zhang F, Kang Y, Wang M, et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet. 2018 Nov15;27(22):3936–3950.
- Zhou KI, Parisien M, Dai Q, et al. N(6)-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol. 2016 Feb 27;428(5):822–833.
- Liang Z, Riaz A, Chachar S, et al. Epigenetic modifications of mRNA and DNA in Plants. Mol Plant. 2020 Jan 6;13(1):14–30.
- Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016 Sep 19;44(16):7511–7526.
- Topisirovic I, Svitkin YV, Sonenberg N, et al. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):277–298.
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016 Feb 25;530(7591):441–446.
- Li X, Xiong X, Wang K, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016 May;12(5):311–316.
- Liu F, Clark W, Luo G, et al. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016 Oct 20;167(3):816–828.e16.
- Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 2016 Dec 29;14(1):23–31.
- Linder B, Grozhik AV, Olarerin-George AO, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015 Aug;12(8):767–772.
- Boulias K, Toczydłowska-Socha D, Hawley BR, et al. Identification of the m(6)Am methyltransferase PCIF1 Reveals the Location and Functions of m(6)Am in the Transcriptome. Mol Cell. 2019 Aug 8;75(3):631–643.e8.
- Sendinc E, Valle-Garcia D, Dhall A, et al. PCIF1 Catalyzes m6Am mRNA Methylation to Regulate Gene Expression. Mol Cell. 2019 Aug 8;75(3):620–630.e9.
- Jones JD, Monroe J, Koutmou KS. A molecular-level perspective on the frequency, distribution, and consequences of messenger RNA modifications. Wiley Interdiscip Rev RNA. 2020 Jul;11(4):e1586.
- Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012 Jun;40(11):5023–5033.
- Amort T, Rieder D, Wille A, et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017 Jan 5;18(1):1.
- Huang T, Chen W, Liu J, et al. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol. 2019 May;26(5):380–388.
- Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 2019 Jan 30;10:2.
- Selmi T, Hussain S, Dietmann S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021 Jan 25;49(2):1006–1022.
- Menezes MR, Balzeau J, Hagan JP. 3ʹ RNA uridylation in epitranscriptomics, gene regulation, and disease. Front Mol Biosci. 2018;5:61.
- Spenkuch F, Motorin Y, Helm M. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol. 2014;11(12):1540–1554.
- Carlile TM, Rojas-Duran MF, Gilbert WV. Pseudo-Seq: genome-wide detection of pseudouridine modifications in RNA. Methods Enzymol. 2015;560:219–245.
- Schwartz S, Bernstein DA, Mumbach MR, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014 Sep 25;159(1):148–162.
- Li X, Zhu P, Ma S, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015 Aug;11(8):592–597.
- Eukaryotic KM. RNA 5ʹ-End NAD(+) Capping and DeNADding. Trends Cell Biol. 2018 Jun;28(6):454–464.
- Peng X, Xu X, Wang Y, et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell. 2018 May 14;33(5):817–828.e7.
- Reily C, Stewart TJ, Renfrow MB, et al. Glycosylation in health and disease. Nat Rev Nephrol. 2019 Jun;15(6):346–366.
- Okada N, Shindo-Okada N, Nishimura S. Isolation of mammalian tRNAAsp and tRNATyr by lectin-Sepharose affinity column chromatography. Nucleic Acids Res. 1977 Feb;4(2):415–423.
- Flynn RA, Pedram K, Malaker SA, et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell. 2021 Jun 10;184(12):3109–3124.e22.
- Anderson SJ, Kramer MC, Gosai SJ, et al. N(6)-Methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep. 2018 Oct 30;25(5):1146–1157.e3.
- Zhang F, Zhang YC, Liao JY, et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019 May;15(5):e1008120.
- Cui X, Liang Z, Shen L, et al. 5-Methylcytosine RNA methylation in Arabidopsis thaliana. Mol Plant. 2017 Nov6;10(11):1387–1399.
- David R, Burgess A, Parker B, et al. Transcriptome-Wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and Noncoding RNAs. Plant Cell. 2017 Mar;29(3):445–460.
- Zhong S, Li H, Bodi Z, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008 May;20(5):1278–1288.
- Shen L, Liang Z, Gu X, et al. N(6)-Methyladenosine RNA modification regulates shoot stem cell fate in arabidopsis. Dev Cell. 2016 Jul 25;38(2):186–200.
- Shen L, Liang Z, Yu H. Dot blot analysis of N6-methyladenosine RNA modification levels. Bio-protocol. 2017;7(1):e2095. 2017/01/05.
- Rajecka V, Skalicky T, Vanacova S. The role of RNA adenosine demethylases in the control of gene expression. BBA Gene Regul Mech. 2019 Mar;1862(3):343–355.
- Liang Z, Geng Y, and Ji C, et al. Mesostigma viride genome and transcriptome provide insights into the origin and evolution of streptophyta. Adv Sci. 2020;71:1901850.
- Růžička K, Zhang M, Campilho A, et al. Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017 Jul;215(1):157–172.
- Duan HC, Wei LH, Zhang C, et al. ALKBH10B Is an RNA N(6)-Methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell. 2017 Dec;29(12):2995–3011.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281–297.
- Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019 Jan;20(1):21–37.
- O’Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136(2):215–233.
- Broughton JP, Lovci MT, Huang JL, et al. Pairing beyond the Seed Supports MicroRNA targeting specificity. Mol Cell. 2016 Oct 20;64(2):320–333.
- Makarova JA, Shkurnikov MU, Wicklein D, et al. Intracellular and extracellular microRNA: an update on localization and biological role. Prog Histochem Cytochem. 2016 Nov;51(3–4):33–49.
- Pu M, Chen J, Tao Z, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019 Feb;76(3):441–451.
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017 Mar;16(3):203–222.
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3ʹ UTRs and near stop codons. Cell. 2012 Jun 22;149(7):1635–1646.
- Han J, Wang JZ, Yang X, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019 Jun 22;18(1):110.
- Peng W, Li J, Chen R, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019 Sep 6;38(1):393.
- Wang H, Deng Q, Lv Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019 Dec 10;18(1):181.
- Zhang J, Bai R, Li M, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019 Apr 23;10(1):1858.
- Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017 Feb;65(2):529–543.
- Wang J, Ishfaq M, Xu L, et al. METTL3/m(6)A/miRNA-873-5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating keap1/Nrf2 pathway. Front Pharmacol. 2019;10:517.
- Yan G, Yuan Y, He M, et al. m(6)A Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol Ther Nucleic Acids. 2020 Mar 6;19:421–436.
- Konno M, Koseki J, Asai A, et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat Commun. 2019 Aug 29;10(1):3888.
- Mi B, Xiong Y, Yan C, et al. Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J Cell Mol Med. 2020 Jun;24(11):6385–6396.
- Yang Z, Li J, Feng G, et al. MicroRNA-145 Modulates N(6)-Methyladenosine Levels by Targeting the 3ʹ-Untranslated mRNA Region of the N(6)-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017 Mar 3;292(9):3614–3623.
- Cui X, Wang Z, Li J, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020 Mar;53(3):e12768.
- Cai X, Wang X, Cao C, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018 Feb 28;415:11–19.
- Gutschner T, Hämmerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl). 2013 Jul;91(7):791–801.
- Li X, Xiong X, Zhang M, et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell. 2017 Dec 7;68(5):993–1005.e9.
- Cheray M, Etcheverry A, Jacques C, et al. Cytosine methylation of mature microRNAs inhibits their functions and is associated with poor prognosis in glioblastoma multiforme. Mol Cancer. 2020 Feb 25;19(1):36.
- Zhu DQ, Lou YF, He ZG, et al. Nucleotidyl transferase TUT1 inhibits lipogenesis in osteosarcoma cells through regulation of microRNA-24 and microRNA-29a. Tumour Biol. 2014 Dec;35(12):11829–11835.
- Thornton JE, Chang HM, Piskounova E, et al. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA. 2012 Oct;18(10):1875–1885.
- Alajez NM, Shi W, Wong D, et al. Lin28b promotes head and neck cancer progression via modulation of the insulin-like growth factor survival pathway. Oncotarget. 2012 Dec;3(12):1641–1652.
- Cai WY, Wei TZ, Luo QC, et al. The Wnt-β-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci. 2013 Jul 1;126(Pt 13):2877–2889.
- Ustianenko D, Hrossova D, Potesil D, et al. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013 Dec;19(12):1632–1638.
- Hunter RW, Liu Y, Manjunath H, et al. Loss of Dis3l2 partially phenocopies Perlman syndrome in mice and results in up-regulation of Igf2 in nephron progenitor cells. Genes Dev. 2018 Jul 1;32(13–14):903–908.
- Song J, Zhuang Y, Zhu C, et al. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol. 2020 Feb;16(2):160–169.
- Xhemalce B, Robson SC, Human KT. RNA methyltransferase BCDIN3D regulates microRNA processing. Cell. 2012 Oct 12;151(2):278–288.
- Jiao X, Doamekpor SK, Bird JG, et al. 5ʹ End nicotinamide adenine dinucleotide cap in human cells promotes RNA Decay through DXO-Mediated deNADding. Cell. 2017 Mar 9;168(6):1015–1027.e10.
- Walters RW, Matheny T, Mizoue LS, et al. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2017 Jan 17 114(3):480–485.
- Wang J, Mei J, Ren G. Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci. 2019;10:360.
- Zhao Y, Yu Y, Zhai J, et al. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012 Apr 24;22(8):689–694.
- Wang X, Zhang S, Dou Y, et al. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3ʹ tailing of small RNAs in Arabidopsis. PLoS Genet. 2015 Apr;11(4):e1005091.
- Tu B, Liu L, Xu C, et al. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 2015 Apr;11(4):e1005119.
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science (New York, NY). 2008 Sep 12;321(5895):1490–1492.
- Yu Y, Ji L, Le BH, et al. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017 Feb;15(2):e2001272.
- Ibrahim F, Rymarquis LA, Kim EJ, et al. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010 Feb 23 107(8):3906–3911.
- Liang H, Jiao Z, Rong W, et al. 3ʹ-Terminal 2ʹ-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic Acids Res. 2020 Jul 27;48(13):7027–7040.
- Yang A, Bofill-De Ros X, Shao TJ, et al. 3ʹ uridylation confers miRNAs with non-canonical target repertoires. Mol Cell. 2019 Aug 8;75(3):511–522.e4.
- Zeng J, Gupta VK, Jiang Y, et al. Cross-kingdom small RNAs among animals, plants and microbes. Cells. 2019 Apr 23;8(4):371.
- Baier SR, Nguyen C, Xie F, et al. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers [Article]. J Nutr. 2014;144(10):1495–1500.
- Zhang L, Hou D, Chen X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012 Jan;22(1):107–126.
- Dávalos A, Henriques R, and Latasa MJ, et al. Literature review of baseline information on non-coding RNA (ncRNA) to support the risk assessment of ncRNA-based genetically modified plants for food and feed.EFSA supporting publication . 2019;16(8):1688E.
- Del Pozo-Acebo L, López de Las Hazas MC, Margollés A, et al. Eating microRNAs: pharmacological opportunities for cross-kingdom regulation and implications in host gene and gut microbiota modulation. Br J Pharmacol. 2021 Jun;178(11):2218–2245.
- Tomé-Carneiro J, Fernández-Alonso N, Tomás-Zapico C, et al. Breast milk microRNAs harsh journey towards potential effects in infant development and maturation. Lipid encapsulation can help. Pharmacol Res. 2018;132:21–32.
- Samad AFA, Kamaroddin MF, Cross-Kingdom SM. Regulation by plant microRNAs provides novel insight into gene regulation. Advances in Nutrition (Bethesda, Md). 2021 Feb 1;12(1):197–211.
- Yang J, Elbaz-Younes I, Primo C, et al. Intestinal permeability, digestive stability and oral bioavailability of dietary small RNAs. Sci Rep. 2018 Jul 6;8(1):10253.
- Wang X, Ren X, Ning L, et al. Stability and absorption mechanism of typical plant miRNAs in an in vitro gastrointestinal environment: basis for their cross-kingdom nutritional effects. J Nutr Biochem. 2020 Jul;81:108376.
- Luo Y, Wang P, Wang X, et al. Detection of dietetically absorbed maize-derived microRNAs in pigs. Sci Rep. 2017 Apr 5;7(1):645.
- Jung M, Schaefer A, Steiner I, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010 Jun;56(6):998–1006.
- Zhou Z, Li X, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015 Jan;25(1):39–49.
- Huang H, Cd D, Wang TTY. Extensive degradation and low bioavailability of orally consumed corn miRNAs in mice. Nutrients. 2018 Feb 15;10(2):215.
- Dickinson B, Zhang Y, Petrick JS, et al. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013 Nov;31(11):965–967.
- Howard KM, Jati Kusuma R, Baier SR, et al. Loss of miRNAs during processing and storage of cow’s (Bos Taurus) milk. J Agric Food Chem. 2015 Jan 21;63(2):588–592.
- Micó V, Martín R, Lasunción MA, et al. Unsuccessful detection of plant microRNAs in beer, extra virgin olive oil and human plasma after an acute ingestion of extra virgin olive oil. Plant Foods Hum Nutr. 2016 Mar;71(1):102–108.
- Dickinson B, Zhang Y, Petrick J S, Heck G, Ivashuta S and Marshall W S. (2013). Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol, 31(11), 965–967. 10.1038/nbt.2737
- Title A C, Denzler R and Stoffel M. (2015). Uptake and Function Studies of Maternal Milk-derived MicroRNAs. Journal of Biological Chemistry, 290(39), 23680–23691. 10.1074/jbc.M115.676734
- Chen Q, Zhang F, Dong L, et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2021 Mar;31(3):247–258.
- Mullokandov G, Baccarini A, Ruzo A, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012 Jul 1;9(8):840–846.
- Seitz H. Redefining microRNA targets. Curr Biol. 2009 May 26;19(10):870–873.
- Brown BD, Gentner B, Cantore A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007 Dec;25(12):1457–1467.
- Park JH, Shin SY, Shin C. Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res. 2017 Feb 28;45(4):1569–1583.
- Mellis D, Caporali A. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem Soc Trans. 2018 Feb 19;46(1):11–21.
- Shah MY, Ferrajoli A, Sood AK, et al. microRNA therapeutics in cancer - an emerging concept. EBioMedicine. 2016 Oct;12:34–42.
- Jones MR, Quinton LJ, Blahna MT, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009 Sep;11(9):1157–1163.
- Adachi H, Hengesbach M, Yu YT, et al. From antisense RNA to RNA modification: therapeutic potential of RNA-Based technologies. Biomedicines. 2021 May 14;9(5):550.
- Zha X, Xi X, Fan X, et al. Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. Aging (Albany NY). 2020 May 4;12(9):8137–8150.
- Zhu Y, Pan X, Du N, et al. ASIC1a regulates miR-350/SPRY2 by N(6) -methyladenosine to promote liver fibrosis. FASEB J. 2020 Nov;34(11):14371–14388.
- Chen J, Wang H, Yang X, et al. Consumption of miRNA-mediated insect-resistant transgenic rice pollen does not harm Apis mellifera adults. J Agric Food Chem. 2021 Apr 14;69(14):4234–4242.
