?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
This study investigated the expression of novel mutations of the β2-microglobulin (B2M) gene in bovine genomic DNA isolated from milk-borne leukocytes and tested the association of these mutations with milk quality traits in Holstein cows. DNA was extracted using a novel protocol allowing for the amplification of long fragments of DNA isolated from small amounts of unprocessed milk samples. Genomic mutations at the B2M locus were explored by DNA sequencing using specific primers and a cluster of five identified within a 729-bp region of intron 3. Specifically, mutations at 7008(G>T), 7202(A>G), 7432(G>T) and 7437(G>C) were all found to be significantly associated with milk protein percentage (P < 0.05), and a mutation at 7026(G>A) was significantly associated with somatic cell count (P < 0.05). The potential to use these mutations as further tools for the pre-evaluation of milk quality is discussed.
RESUMEN
El presente estudio investigó cómo se manifiestan mutaciones novedosas del gen β2-microglobulina en el ADN genómico bovino aislado de leucocitos transmitidos en la leche, a la vez que sometió a prueba la asociación de dichas mutaciones con las características de calidad de la leche obtenida de vacas Holstein. El ADN se extrajo utilizando un protocolo innovador que permite amplificar largos fragmentos del ADN aislados de pequeñas cantidades de muestras de leche sin procesar. Se examinaron las mutaciones genómicas ocurridas en el locus del (B2M) mediante la secuenciación del ADN, utilizando para ello partidores específicos y un clúster de cinco, identificados en una región de 729 bp del intrón 3. Específicamente, se determinó que las mutaciones en 7008 (G>T), 7202 (A>G), 7432 (G>T) y 7437 (G>C) se encuentran significativamente asociadas con el porcentaje de proteína láctea (MPP) (P<0,05), y que una mutación en 7026 (G>A) se asocia de manera significativa con el conteo de células somáticas (SCC) (P<0,05). Para finalizar, se discute la posibilidad de usar estas mutaciones como herramientas adicionales para la preevaluación de la calidad de la leche.
PALABRAS CLAVE:
1. Introduction
Increasing concerns about food safety generally and the impact of dairy products on human health specifically have resulted in tightening regulatory legislation and targeted supplier payment schemes based on the quality of milk composition (Friggens & Rasmussen, Citation2001). These concerns also provide a strong impetus to improve milk quality traits, such as milk yield (MY), milk fat percentage (MFP), milk protein percentage (MPP) and somatic cell count (SCC), each of which has been considered as an important determinant of product quality and resultant consumer satisfaction (VanRaden, Citation2006). Given this landscape, the development of increasingly sophisticated means for predicting and assessing milk composition would be expected to promote the growth of the dairy industry.
β2-microglobulin (B2M) is a 12-kD secreted protein which has crucial roles in a broad range of biological processes, notably immune modulation (Tin et al., Citation2013), melanocyte stimulation (Hofmann, Kiecker, Küchler, Kors, & Trefzer, Citation2011) and kidney function, and which contribute to the progression of cardiovascular diseases (Bash, Astor, & Coresh, Citation2010; Matsushita et al., Citation2010), peripheral arterial disease (Wilson et al., Citation2007), multiple myeloma (Rossi et al., Citation2010) and inflammatory diseases (Bianchi et al., Citation2001; Zissis et al., Citation2001). Genetic ablation in mice results in aberrant-immune cell composition, a phenotype which resembles that of humans who carry rare mutations of the B2M gene (Crump, Grusby, Glimcher, & Cantor, Citation1993; Liao, Bix, Zijlstra, Jaenisch, & Raulet, Citation1991; Smithies & Poulik, Citation1972). In beef cattle, 12 mutations in the regions of exon 2 and 4 of the B2M gene have previously been described and found to be associated with changes of milk protein and variable levels of serum immunoglobulin G (IgG) contributing to elevated SCC (Clawson et al., Citation2004; Sanchez et al., Citation2004; Worley et al., Citation2009).
This strong association to immune-regulatory function makes the B2M gene a particularly attractive candidate marker of milk quality. We have recently developed methods for efficient isolation of bovine genomic DNA directly from unprocessed milk samples, as an alternative to sampling blood which causes significant stress responses in cows (Liu, Gao, Yang, Ku, & Zan, Citation2014). In the current work, bovine genomic DNA extracted from milk was used to screen for potential mutations in the B2M gene of Holstein dairy cows by direct sequencing. Further, the relationship of these mutations to milk quality traits was examined. A total of five mutations were discovered and shown to be variably associated with key milk quality traits, including MY, MFP, MPP and SCC. As such, they may provide further tools for the assessment of measurable differences in milk quality traits in these herds.
2. Material and methods
2.1. Samples source and milk quality traits measurement
Raw milk samples (15 ml) were collected at the first milking of the day from 79 unrelated and randomly selected Chinese Holstein cows, all about 3 years old and in a single herd. Once collected, the samples were transported on ice packs and subsequently maintained at optimum low temperature prior to analysis. The milk quality traits MY, MFP, MPP and SCC were measured using standard protocols on a Foss MilkoScan FT120 instrument (Foss Electric, DK-3400 Hillerød, Denmark). Each milk quality trait was measured by a single investigator, in order to minimize measurement bias. Values were represented with mean ± standard error of the mean. Then, DNA was extracted from leukocytes isolated from the milk samples using a novel method that we have previously reported and based on long-fragment DNA amplification from small amounts of raw milk (Liu et al., Citation2014).
2.2. Detection of mutations and genotyping
Based on sequence of bovine B2M gene (GenBank accession No. NC_007308.5), one pair of primers (5′-GGC TTT CCC AGC ATC ACT AAC-3′ and 5′-TCA CAG CAC CAC CAA ACT TAT CT-3′) was designed to amplify a 729-bp product of the B2M intron 3. Polymerase chain reaction (PCR) was conducted in 30 μl reaction mixtures containing 50 ng DNA templates, 10 pM of each primer, 0.20 mM dNTP, 2.5 mM MgCl2 and 0.5 U Taq DNA polymerase (TaKaRa, Dalian, PR China). The PCR protocol consisted of 95°C for 5 min followed by 30 cycles of 94°C for 30 s, 60°C annealing for 30 s and 72°C for 30 s, and a final extension at 72°C for 10 min. The end products were purified with a Wizard Prep PCR purification kit (Shanghai Bioasia Biotechnology, PR China) and sequenced using established protocols (Beijing Aolaibo Biotechnology, PR China; Applied Biosystems 3730xl DNA Sequencer, Foster City, CA, USA). Mutations in the B2M gene were identified across 79 different milk samples and compared with one another using SeqMan software (DNASTAR, Inc., Madison, WI, USA).
2.3. Statistic analysis
While a substantial measure of the variance in milk quality traits can be attributed to factors such as genotype, age, breed and herd status, and geographic location, the drivers of the remaining variability remain poorly defined. Since individual cow’s milk was collected, two factors (genotype and other random errors) were involved in the statistical model while the effects of cow factors and location were excluded during experimental design.
Following published approaches (Nei & Li, Citation1979; Nei & Roychoudhury, Citation1974), the established bioinformatic parameters for “proportions of genotypes”, allelic frequencies, gene heterozygosity and polymorphism information content (PIC) were statistically analyzed. Heterozygosity and PIC were used to indicate the extent of genetic variation at the B2M gene locus. The association between genotypes of mutation in B2M gene and the milk quality traits (MY, MFP, MPP and SCC) was analyzed by mathematical method and SAS software (SAS Institute Inc., Cary, NC, USA). A statistical linear model obtained was as follows:
where Yi is the measured milk quality trait, µ is the overall mean for each trait, Gi is the genotype effect and εj is the random errors effect.
For the haplotype analyses, the model was subjected to the same covariates in a similar manner and the degree of linkage disequilibrium (LD), reflecting the nonrandom association of alleles at different loci, was investigated. Currently, the most commonly used parameters to determine LD are D′ and r2, where D′ is a measure of LD relative to the maximum possible value (given the allelelic frequency of the mutations) and was calculated from the frequencies of the haplotype of the mutation pairs. Meanwhile, r2 is the square of correlation between pairs of mutations and can be used as a standardization measurement of LD between alleles existing at two loci. These two parameters of LD were measured using Haploview 2.05 software (Barrett, Fry, Maller, & Daly, Citation2005; Gabriel et al., Citation2002) after the haplotypes and their frequencies were inferred using a published algorithm (Stephens, Smith, & Donnelly, Citation2001). The phase probabilities of all the polymorphic sites for the haplotypes were calculated using PHASE software (http://depts.washington.edu/ventures/UW_Technology/Express_Licenses/PHASEv2.php).
3. Results
3.1. Determination of mutations in B2M gene
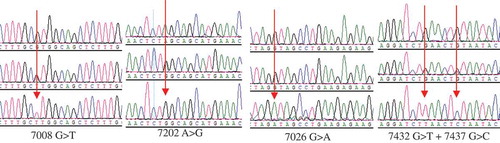
By direct DNA sequencing, five discrete mutations were identified in intron 3 of the B2M gene. Denoted as 7008G>T, 7026G>A, 7202A>G, 7432G>T and 7437G>C, they were all identified within a 729-bp PCR product, whose DNA sequencing maps are shown in .
3.2. Analysis of bioinformatics parameters and association between mutations and milk quality traits
Within the population of 79 animals, we further analyzed the proportions of genotypes, allelic frequencies, gene heterozygosity and PIC of mutations, as summarized in . These data suggest that the identified mutations contribute to three separable genotypes representing the “common allele”, “heterozygotes” and “homozygotes” for the rare allele, respectively. The GT genotype on 7008G>T mutation contributed 0.6076 to this variability and representing more than half of the effect of three genotypes, and almost twice the proportion attributable to CC genotype. The GG genotype on the 7026G>A mutation, the AG genotype on the 7202A>G mutation and the GG genotype on the 7432G>T respectively contributed 0.4684, 0.6456 and 0.5696 to the variability, which was almost twice the proportion attributable to the GA, GG and TT genotype, respectively. The GG genotype on the 7437G>C mutation contributed 0.6203 to the variability, which was almost three times the proportion attributable to the CC genotype. As to the allelic frequencies of the 5 mutations, the frequencies of the normal allele ranged from 0.5823 to 0.7089. Meanwhile, the ranges (from mutation sites 7437G>C to 7026G>A) of heterozygosis and PIC of the 5 mutations were from 0.4128 to 0.4865 and from 0.3276 to 0.3681, respectively, and there was no significant change in them.
Table 1. Proportions of genotypes, allele frequencies, gene heterozygosity and PIC indexes of intron 3 of B2M gene in 79 different milk samples.
Tabla 1. Proporciones de genotipos, frecuencia de alelos, heterozigosidad de genes e índices PIC del intrón 3 del gen B2M en 79 muestras diferentes de leche.
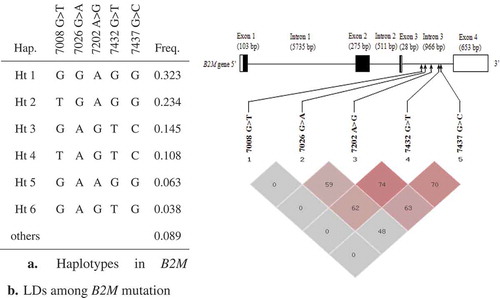
Theoretically, there are 32 (25) possible haplotypes for the 5 mutations in the bovine B2M gene, although we only identified 6 genotyped mutations (GGAGG, TGAGG, GAGTC, TAGTC, GAAGG and GAGTG). Four common haplotypes (freq. > 0.1) were constructed and they also showed strong LDs (). According to LD analysis (), there was a tight LD (r2 > 0.33) between each of the mutations, except for mutation 7008G>T which presented a weaker LD (r2 < 0.33) with all the other mutations. Therefore, the five mutations were chosen as “tag mutations” for analysis of their associations with milk quality traits.
Figure 2. Gene haplotype and LD coefficients in B2M. (a) Haplotypes of B2M. Haplotypes with frequency >0.03 are presented. Others contain rare haplotypes. (b) LD parameters (including D′ and r2) among B2M mutations. The numbers are r2 value (%).
Figura 2. Haplotipo de gen y coeficientes LD en B2M. (a) Haplotipos de B2M. Se presentan los haplotipos con una frecuencia >0,03. Los otros contienen haplotipos escasos. (b) Parámetros LD (incluyendo D’ y r2) entre mutaciones del B2M. Los números corresponden a valores r2 (%).
Meanwhile, the relationship between the genotypes of the 79 subject cows and the 4 milk quality traits was analyzed (phenotypic data not shown) and the associations summarized in . Notably, the mutation 7008G>T had a significant relationship with MPP (P < 0.05) and those samples with the genotype TT had a lower mean value for MPP than those with the GG and GT genotype (P = 0.0237). The mutation 7202A>G also had a significant relationship with MPP (P < 0.05) and samples with the genotype AG and GG had a lower mean value of MPP than those with the AA genotype (P = 0.0138). Again, the mutation 7432G>T had a significant relationship with MPP (P < 0.05) and samples with the genotype GT had a lower mean value of MPP than those with the GG genotype (P = 0.0446). Finally, the mutation 7437G>C also had a significant relationship with MPP (P < 0.05) and samples with the genotype GC had a lower mean value of MPP than those with the GG genotype (P = 0.0106). Additionally, the mutation 7026G>A had a significant relationship with SCC (P < 0.05) and those samples with the genotype AA had a lower mean value of SCC than those with the GA genotype (P = 0.0484).
Table 2. Association of B2M mutation with milk quality traits on 79 milk samples.
Tabla 2. Asociación de la mutación del B2M con las características de calidad de la leche en 79 muestras de leche.
4. Discussion
We have identified five mutations in the B2M gene of dairy cows which display variable heterozygosity, frequency and PIC score, although the individual variances were relatively small (). In genetic linkage analysis, the PIC value was usually used to detect a specific mutation and a polymorphism within a population, and it depended on the number of available allele and frequency distribution (Wenzl et al., Citation2004). Originally defined as a codominant marker in a linkage study of a rare dominant disease, PIC has more recently been shown to be relevant regardless of the mode of phenotype inheritance (Shete, Tiwari, & Elston, Citation2000). Quantitatively, the degree of polymorphism reflects both the degree of heterozygosity and PIC value and classified as low (PIC < 0.25), median (0.25 < PIC < 0.5) or high (PIC > 0.5), with most populations being low or moderate (Mateescu et al., Citation2005). The PIC values in the present study ranged from 0.3276 to 0.3681, indicating largely moderate degrees of polymorphism in these particular populations The average PIC value of 0.3479 found in our study indicates that, on average, one in three of the intron-derived mutations are expected to be polymorphic between any two native Chinese cattle breeds.
By definition, value of r2 > 0.33 defines strong LD, a measure of the nonrandom association of mutations at different loci in a population (Ardlie, Kruglyak, & Seielstad, Citation2002). Our analysis of the B2M mutations indicates that four of the five mutations were functionally a unit (the exception being the 7008G>T mutation), such that each could serve as a tag mutation, and every pair of mutations with the unit was having a strong LD (r2 > 0.33). Therefore, the single mutation 7008G>T and each one of the other four mutations in the unit could potentially be used as tag mutations to analyze association.
The five mutations we have described were found to have significant associations with MPP and SCC in Holstein dairy (), consistent with a number of recent studies (from across species) which linked functional mutations in B2M gene with impacts on milk composition in the mammary gland of lactating animals, and immune modulation in humans (Crump et al., Citation1993; Liao et al., Citation1991). Further, genome-wide association study (GWAS) identify a mutation of the human B2M gene (rs2255235, P = 3.1 × 10−5) with adult immunity (Wade et al., Citation2011), while a further six mutations, rs9260489 (P = 1.9 × 10−8), rs2023472 (P = 1.0 × 10−4), rs9264638 (P = 1.5 × 10−9), rs2523608 (P = 1.7 × 10−8), rs16899524 (P = 3.8 × 10−6) and rs2596466 (P = 2.2 × 10−3), are found to be associated with immunity-related phenotypes across large human populations (Tin et al., Citation2013). In addition, rare mutations in both mice and sheep B2M gene cause increases in the level of IgG, underpinning higher milk protein content and SCC (Crump et al., Citation1993; Liao et al., Citation1991; Mayer et al., Citation2002). Going beyond simple association, the functional expression of the B2M gene in murine models has been shown to influence more broadly the composition of milk (Cianga, Medesan, Richardson, Ghetie, & Ward, Citation1999). Thus, while a number of studies report the association of B2M gene variants with immune function in animal models and human populations, there are still very little data relating such mutations to milk quality traits in dairy herd populations, in which the biology of milk composition and immune status are of paramount importance as determinants of food safety in a key worldwide human supply chain. Our study is a preliminary step in providing the essential tools for analyzing mutations and genetic effects at the bovine B2M gene locus.
5. Conclusion
In summary, we identified multiple mutations in intron 3 of the B2M immune-related gene using DNA isolated from milk samples and investigated its association with milk quality traits across a population of Holstein dairy cows. Analysis of the proportion of genotypes, allelic frequencies, heterozygosity and PIC was performed to explain the mutation distribution across the population. Association analysis revealed that mutations including 7008G>T, 7202A>G, 7432G>T and 7437G>C were all significantly associated with MPP (P = 0.0237, 0.0138, 0.0446 and 0.0106, respectively) and that 7026G>A was significantly associated with SCC (P = 0.0484). Thus, these mutations at the B2M locus should be useful for pre-evaluation and selection for of milk quality by genotyping. The enhanced screening of raw milk for quality traits is required to meet increasing expectations of food safety and should translate to increased economic benefit for producers.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ardlie, K. G., Kruglyak, L., & Seielstad, M. (2002). Patterns of linkage disequilibrium in the human genome. Nature Reviews Genetics, 3, 299–309. doi:10.1038/nrg777
- Barrett, J. C., Fry, B., Maller, J., & Daly, M. J. (2005). Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England), 21, 263–265. doi:10.1093/bioinformatics/bth457
- Bash, L. D., Astor, B. C., & Coresh, J. (2010). Risk of incident ESRD: A comprehensive look at cardiovascular risk factors and 17 years of follow-up in the atherosclerosis risk in communities (ARIC) study. American Journal of Kidney Diseases, 55, 31–41. doi:10.1053/j.ajkd.2009.09.006
- Bianchi, C., Donadio, C., Tramonti, G., Consani, C., Lorusso, P., & Rossi, G. (2001). Reappraisal of serum beta2-microglobulin as marker of GFR. Renal Failure, 23, 419–429. doi:10.1081/JDI-100104725
- Cianga, P., Medesan, C., Richardson, J. A., Ghetie, V., & Ward, E. S. (1999). Identification and function of neonatal Fc receptor in mammary gland of lactating mice. European Journal of Immunology, 29, 2515–2523. doi:10.1002/(ISSN)1521-4141
- Clawson, M. L., Heaton, M. P., Chitko-McKown, C. G., Fox, J. M., Smith, T. P. L., Snelling, W. M., … Laegreid, W. W. (2004). Beta-2-microglobulin haplotypes in U.S. beef cattle and association with failure of passive transfer in newborn calves. Mammalian Genome, 15, 227–236. doi:10.1007/s00335-003-2320-x
- Crump, A. L., Grusby, M. J., Glimcher, L. H., & Cantor, H. (1993). Thymocyte development in major histocompatibility complex-deficient mice: Evidence for stochastic commitment to the CD4 and CD8 lineages. Proceedings of the National Academy of Sciences, 90, 10739–10743. doi:10.1073/pnas.90.22.10739
- Friggens, N. C., & Rasmussen, M. D. (2001). Milk quality assessment in automatic milking systems: Accounting for the effects of variable intervals between milkings on milk composition. Livestock Production Science, 73, 45–54. doi:10.1016/S0301-6226(01)00228-7
- Gabriel, S. B., Schaffner, S. F., Nguyen, H., Moore, J. M., Roy, J., Blumenstie, B., … Altshuler, D. (2002). The structure of haplotype blocks in the human genome. Science, 296, 2225–2229. doi:10.1126/science.1069424
- Hofmann, M. A., Kiecker, F., Küchler, I., Kors, C., & Trefzer, U. (2011). Serum TNF-ɑ, B2M and sIL-2R levels are biological correlates of outcome in adjuvant IFN-ɑ2b treatment of patients with melanoma. Journal of Cancer Research and Clinical Oncology, 137, 455–462. doi:10.1007/s00432-010-0900-1
- Liao, N. S., Bix, M., Zijlstra, M., Jaenisch, R., & Raulet, D. (1991). MHC class I deficiency: Susceptibility to natural killer (NK) cells and impaired NK activity. Science, 253, 199–202. doi:10.1126/science.1853205
- Liu, Y. F., Gao, J. L., Yang, Y. F., Ku, T., & Zan, L. S. (2014). Novel extraction method of genomic DNA suitable for long-fragment amplification from small amounts of milk. Journal of Dairy Science, 97, 6804–6809. doi:10.3168/jds.2014-8066
- Mateescu, R. G., Zhang, Z., Tsai, K., Phavaphutanon, J., Burton-Wurster, N. I., Quaas, R., … Todhunter, R. J. (2005). Analysis of allele fidelity, polymorphic information content, and density of microsatellites in a genome-wide screening for hip dysplasia in a crossbreed pedigree. Journal of Heredity, 96, 847–853. doi:10.1093/jhered/esi109
- Matsushita, K., Van Der, V. M., Astor, B. C., Woodward, M., Levey, A. S., & De Jong, P. E. (2010). Association of estimated glomerular filtration rate and albuminuria with allcause and cardiovascular mortality in general population cohorts: A collaborative meta-analysis. Lancet, 37, 2073–2081.
- Mayer, B., Zolnai, A., Frenyo, L. V., Jancsik, V., Szentirmay, Z., Hammarström, L., & Kacskovics, I. (2002). Redistribution of the sheep neonatal Fc receptor in the mammary gland around the time of parturition in ewes and its localization in the small intestine of neonatal lambs. Immunology, 107, 288–296. doi:10.1046/j.1365-2567.2002.01514.x
- Nei, M., & Li, W. H. (1979). Mathematic model for studying genetic variation in terms of restriction endonucleaes. Proceedings of the National Academy of Sciences, 76, 5269–5273. doi:10.1073/pnas.76.10.5269
- Nei, M., & Roychoudhury, A. K. (1974). Sampling variance of heterozygosity and genetic distance. Genetics, 76, 379–390.
- Rossi, D., Fangazio, M., De Paoli, L., Puma, A., Riccomagno, P., Pinto, V., … Gaidano, G. (2010). Beta-2-microglobulin is an independent predictor of progression in asymptomatic multiple myeloma. Cancer, 116, 2188–2200.
- Sanchez, J., Markham, F., Dohoo, I., Sheppard, J., Keefe, G., & Lesliec, K. (2004). Milk antibodies against Ostertagia ostertagi: Relationships with milk IgG and production parameters in lactating dairy cattle. Veterinary Parasitology, 120, 319–330. doi:10.1016/j.vetpar.2004.01.010
- Shete, S., Tiwari, H., & Elston, R. C. (2000). On estimating the heterozygosity and polymorphism information content value. Theoretical Population Biology, 57, 265–271. doi:10.1006/tpbi.2000.1452
- Smithies, O., & Poulik, M. D. (1972). Dog homologue of human β2-microglobulin. Proceedings of the National Academy of Sciences, 69, 2914–2917. doi:10.1073/pnas.69.10.2914
- Stephens, M., Smith, N. J., & Donnelly, P. (2001). A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics, 68, 978–989. doi:10.1086/319501
- Tin, A., Astor, B. C., Boerwinkle, E., Hoogeveen, R. C., Coresh, J., & Kao, W. H. L. (2013). Genome-wide association study identified the human leukocyte antigen region as a novel locus for plasma beta-2 microglobulin. Human Genetics, 132, 619–627. doi:10.1007/s00439-013-1274-7
- VanRaden, P. M., & Multi-State Project S-1008 (2006). New Merit as a measure of lifetime profit: 2006 Revision, http://aipl.arsusda.gov/reference/nmcalc.htm.
- Wade, R., Di Bernardo, M. C., Rossi, D., Crowther-Swanepoel, D., Gaidano, G., Oscier, D. G., … Houlston, R. S. (2011). Association between single nucleotide polymorphism-genotype and outcome of patients with chronic lymphocytic leukemia in a randomized chemotherapy trial. Haematologica, 96, 1496–1503. doi:10.3324/haematol.2011.043471
- Wenzl, P., Carling, J., Kudrna, D., Jaccoud, D., Huttner, E., & Kleinhofs, A. (2004). Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proceedings of the National Academy of Sciences of the United States of America, 101, 9915–9920. doi:10.1073/pnas.0401076101
- Wilson, A. M., Kimura, E., Harada, R. K., Nair, N., Narasimhan, B., Meng, X. Y., … Cooke, J. P. (2007). β2-microglobulin as a biomarker in peripheral arterial disease: Proteomic profiling and clinical studies. Circulation, 116, 1396–1403. doi:10.1161/CIRCULATIONAHA.106.683722
- Worley, K. C., Gibbs, R. A., Elsik, C. G., Tellam, R. L., Muzny, D. M., Weinstock, G. M., & Adelson, D. L. (2009). The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science, 324, 522–528. doi:10.1126/science.1169588
- Zissis, M., Afroudakis, A., Galanopoulos, G., Palermos, I., Boura, X., Michopoulos, S., & Archimandritis, A. (2001). B2 microglobulin: Is it a reliable marker of activity in inflammatory bowel disease? The American Journal of Gastroenterology, 96, 2177–2183. doi:10.1111/j.1572-0241.2001.03881.x