Figures & data
Figure 2. Agarose gel assays of the DNA protection activity of the six polyphenol monomers. A. resveratrol, B. ruit, C. oxyresveratrol, D. mulberroside A, E. quercetin, F. morin. PBP: phosphate buffered potassium. 1. DNA (3 μL) + PBP (2 μL) + ddH2O (5 μL); 2. DNA (3 μL) + PBP (2 μL) + solvent (5 μL); 3. DNA (3 μL) + PBP (2 μL) + ddH2O (5 μL) + UV (30 min); 4. DNA (3 μL) +PBP (2 μL) + solvent (5 μL) + UV (30 min); 5–11: DNA (3 μL) + PBP (2 μL) + different concentrations of sample solution (5 μL 0.03125 mg/mL to 2.00000 mg/mL) + UV (30 min).
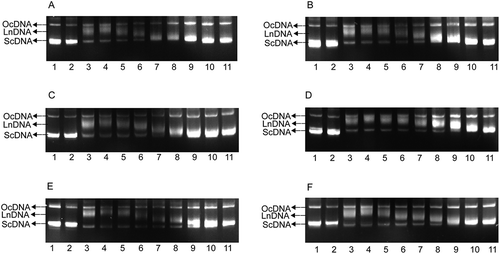
Figure 3. Analysis of ScDNA at the optimum polyphenol concentrations. a Values expressed are means ± S.D. of three parallel measurements. Data in the same column marked with different superscript symbols indicate significant differences (compared with the blank control group of ScDNA; p < 0.01). Re: Resveratrol; Ox: Oxyresveratrol; Ru: Ruit; Mu: Mulberroside A; Qu: Quercetin; Mo: Morin.
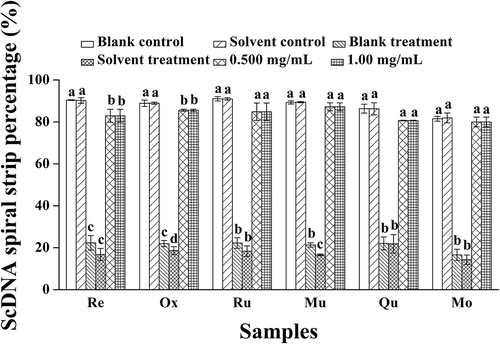
Figure 4. Analysis of OcDNA bands for each experimental group. a Values expressed are means ± S.D. of three parallel measurements. Data in the same column marked with different superscript symbols indicate significant differences (compared with the highest proportion of OcDNA; p < 0.01).Re: Resveratrol; Ox: Oxyresveratrol; Ru: Ruit; Mu: Mulberroside; Qu: Quercetin; Mo: Morin.
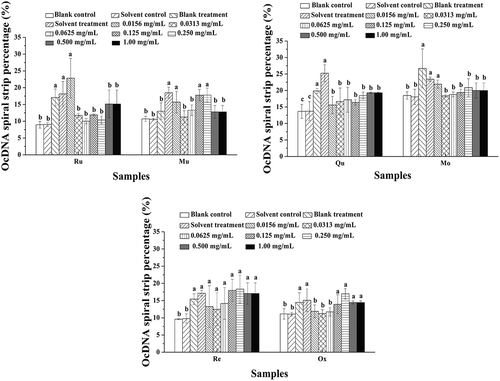
Table 1. The amount of the six polyphenol monomers in Morus alba L. twig extract.a
Figure 5. HPLC chromatograms of the six polyphenol monomers. A. HPLC chromatograms of oxyresveratrol, resveratrol, and mulberroside A standards. a. HPLC chromatograms of oxyresveratrol, resveratrol, and mulberroside A in Morus alba L. twig extract. B. HPLC chromatograms of rutin and quercetin standards. b. HPLC chromatograms of rutin and quercetin in the Morus alba L. twig extract. C. HPLC chromatograms of the morin standard. c. HPLC chromatograms of morin in the Morus alba L. twig extract.
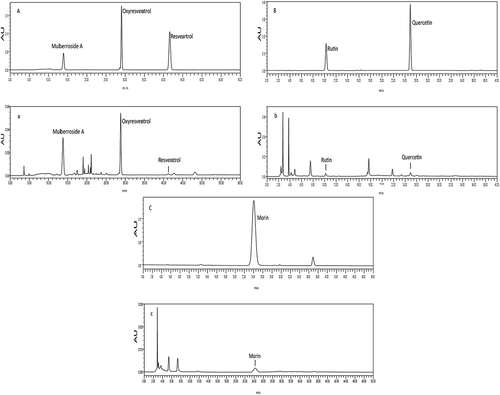
Figure 6. Six standard hybrid system analyses of protection against DNA damage. A. Morus alba L. twig extract; B. mixture of six polyphenol monomers. PBP: phosphate buffered potassium. 1. DNA (3 μL) + PBP (2 μL) + ddH2O (5 μL); 2. DNA (3 μL) + PBP (2 μL) + solvent (5 μL); 3. DNA (3 μL) + PBP (2 μL) + ddH2O (5 μL) + UV (30 min); 4. DNA (3 μL) +PBP (2 μL) + solvent (5 μL) + UV (30 min); A: 5–11: DNA (3 μL) + PBP (2 μL) + different concentrations of Morus alba L. twig extract (5 μL 0.0313 mg/mL to 2.00 mg/mL) + UV (30 min); B: 5–11: DNA (3 μL) + PBP (2 μL) + the mixture of six polyphenol monomers of different concentration of Morus alba L. twig extract (5 μL 0.0313 mg/mL to 2.00 mg/mL) + UV (30 min).

Figure 7. OcDNA band percentage analysis in each experimental group. a Values expressed are means ± S.D. of three parallel measurements. Data in the same column marked with different superscript symbols indicate significant differences (compared with the highest proportion of OcDNA; p < 0.01).
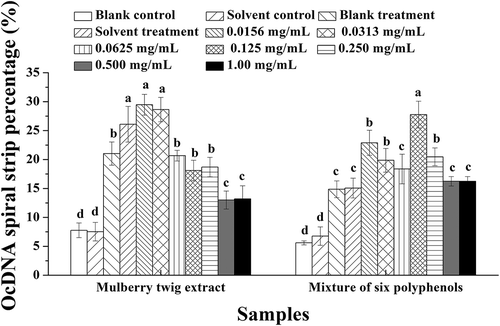
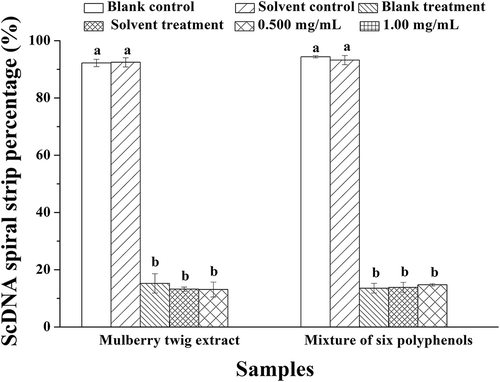
Figure 8. ScDNA band percentage analysis in the presence of optimal polyphenol monomer concentrations. a Values expressed are means ± S.D. of three parallel measurements. Data in the same column marked with different superscript symbols indicate significant differences (compared with the ScDNA of blank control group; p < 0.01).