Figures & data
Table 1. Qualitative phytochemical tests of the JRRAE.
Table 2. Physicochemical characteristics of JRRAE.
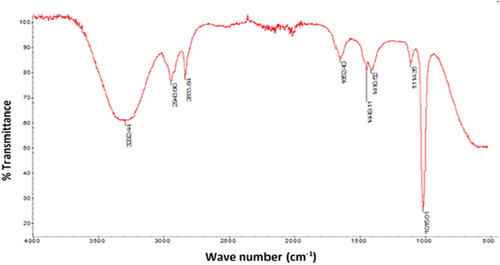
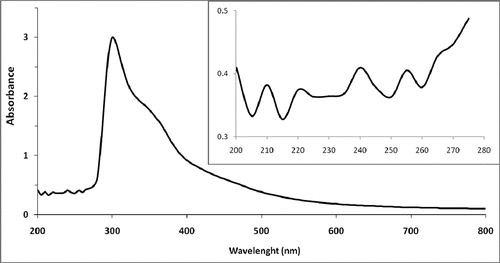
Figure 1b. UV-VIS spectra of JRRAE, HPLC chromatogram of JRRAE: (1) gallic acid, (2) juglone, (3) catechin, (4) vanillic acid, (5) epicatechin, (6) coumarin.IR spectrum of JRRAE.
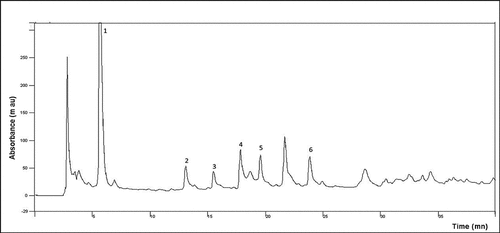
Table 3. Antibacterial activity and effect of JRRAE on pathogenic bacterial adhesion evaluated by the crystal violet test.
Table 4. Bacterial count in the small intestine and feces in control and treated rats for 10 days by JRRAE at 200 mg/kg.
Figure 2. Histopathological effect of the extract on the intestine of S.littoralislarvae (a): Normal appearance. (b): Histopathological aspect of the intestine of S.littoralistreated larvae. V:vesicles formation L:Lysis of epithelial cells. G:Large vacuolation of epithelial cells. Lu:lumen. Ap:apical membrane. Bm:basal membrane. (Magnification × 40).
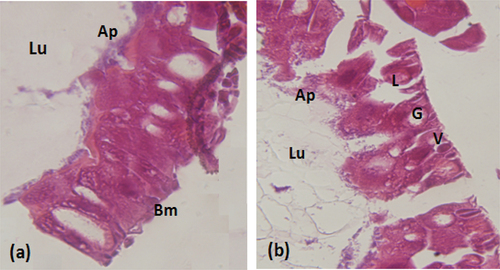
Table 5. Docking binding energies, conventional hydrogen bonding, and the number of closest residues to the docked compounds into the active sites of S. aureus Gyrase (pdb 2 × CT), S. aureus tyrosyl-tRNA synthetase (pdb 1JIJ), human peroxiredoxin 5 (pdb 1HD2), cyclooxygenase-2 (pdb 1C × 2), and trypanothione reductase (pdb 2JK6).
Figure 3. 3D (right) and 2D (left) binding interactions of the stable Coumarin- S. aureus Gyrase, Epicatechin- S. aureus tyrosyl-tRNA synthetase, Juglone- human peroxiredoxin 5, Epicatechin- cyclooxygenase-2, and Epicatechin-trypanothione reductase complexes.
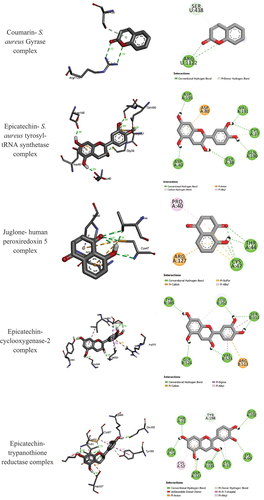
Table 6. ADME properties of the identified compounds.
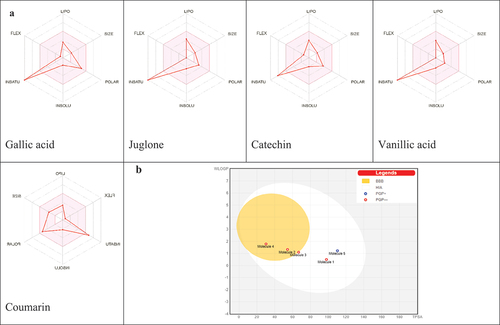
Figure 4. (A) Bioavailability radar of identified compounds based on physicochemical indices ideal for oral bioavailability. LIPO, Lipophilicity: −0.7 < XLOGP3 < þ 5; SIZE, Molecular size: 150 g/mol < mol. wt.<500 g/mol; POLAR, Polarity: 20 Å2 < TPSA<130 Å2; INSOLU, Insolubility: 0 < Log S (ESOL) < 6; INSATU, Insaturation: 0.25 < Fraction Csp3 < 1; FLEX, Flexibility: 0 < Number of rotatable bonds<9. The colored zone is the suitable physicochemical space for oral bioavailability. (B) Boiled-egg (B) model of compounds.