Figures & data
Figure 1. Mean (n = 50) content (µg/g DW) for the analyzed phenolic compounds (A) flavan-3-ols and chlorogenic acids, (B) flavonols in different black currant populations. Error bars indicate ± standard deviation (SD) of means. Codes: 1 = epigallocatechin; 2 = catechin; 3 = epicatechin; 4 = neochlorogenic acid; 5 = chlorogenic acid; 6 = myricetin-malonylglucoside; 7 = quercetin-3-O-galactoside; 8 = quercetin-3-O-glucoside; 9 = quercetin-3-O-rutinoside; 10 = quercetin-3-6-malonylglucoside; 11 = kampferol-3-O-rutinoside; 12 = kaempferol-3-O-glucoside; 13 = isorhamnetin-3-O-rutinoside; 14 = isorhamnetin-3-O-glucoside; 15 = kaempferol-malonylglucoside.
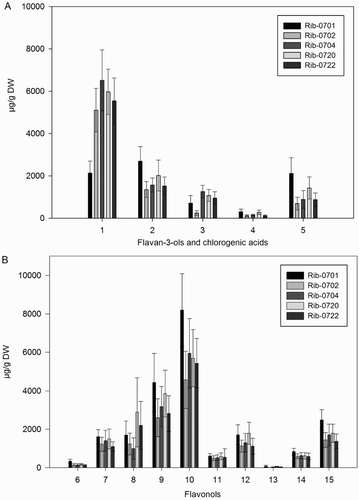
Figure 2. Mean (n = 50) scores (scale 1-9, no disease symptoms to heavy disease symptoms) of foliar diseases between black currant populations. Different letter/letters indicate significant differences (p < .05) between the populations for each foliar disease. Error bars represent ± standard error (SE) of mean.
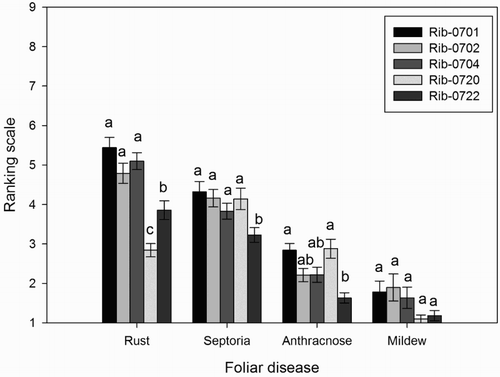
Table 1. Within-groups correlations between discriminating variables and standardized canonical discriminant functions for black currant populations according to their phenolic compounds in leaves.
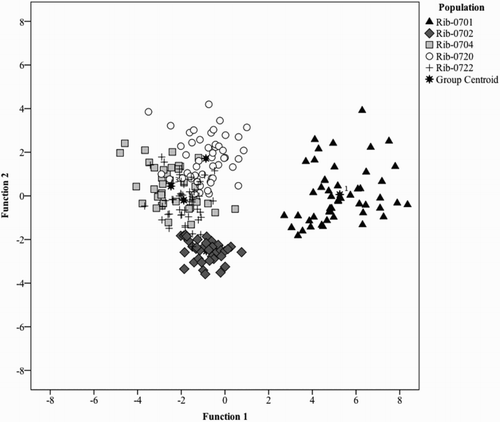
Figure 3. Canonical discriminant analyses plot for scores of phenolic compounds based on the first two canonical discriminant functions separating black currant population.