Figures & data
Figure 1. The conceptualization of an integrative approach. The plasma and brain drug exposure profile are determined by the pharmacokinetics, to drive the target binding and activation of potentially multiple targets. The activation (or inhibition) of these targets elicits multiple downstream biochemical effects, which can be evaluated by proteomics or metabolomics. These processes are described by mathematical expressions as developed in the field of PK/PD modeling.
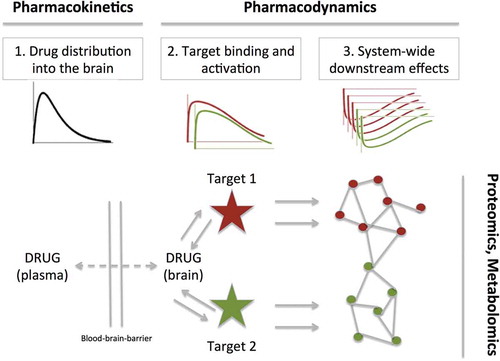
Figure 2. The integrative approach of metabolomics and PK/PD modeling as applied to interspecies scaling in CNS drug development. Such approach starts with animal experiments to collect longitudinal brainECF and plasma samples during treatment with a CNS drug. These samples are analyzed for drug concentrations and metabolomics to subsequently develop a multivariate PK/PD model. By applying the principles of interspecies scaling a humanized model is defined to select doses for the clinical study. Plasma drug concentrations and metabolomics data of the clinical study will be used to recalibrate the model and increase the understanding of interspecies differences.
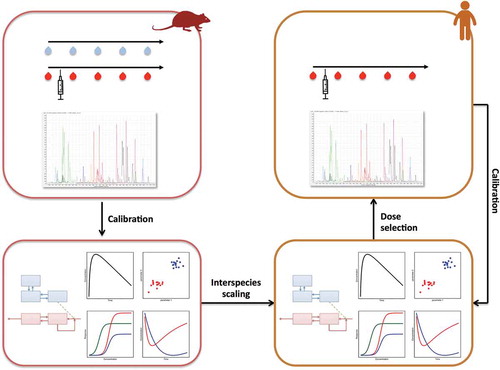
Figure 3. A metabolomics study combined with multivariate PK/PD modeling revealed 6 diverse response patterns (middle) for remoxipride in rats. These response patterns were represented by 18 metabolites that could potentially function as biomarker (right), rendering further validation. The response clusters were associated with 2 known biological pathways (left). Modified from reference [Citation95] with permission of Elsevier.
![Figure 3. A metabolomics study combined with multivariate PK/PD modeling revealed 6 diverse response patterns (middle) for remoxipride in rats. These response patterns were represented by 18 metabolites that could potentially function as biomarker (right), rendering further validation. The response clusters were associated with 2 known biological pathways (left). Modified from reference [Citation95] with permission of Elsevier.](/cms/asset/5f7b7d5a-67c3-4c0d-ab85-da1234a46874/iedc_a_1446935_f0003_oc.jpg)
Figure 4. Brain metabolic phenotypes are reflected in the periphery via three mechanisms: i) individual metabolites distribute to CSF, plasma and urine, and become integrated in the peripheral metabolic phenotype; ii) the brain metabolic phenotype affects the peripheral nervous signaling, thereby controlling the release of peripheral metabolites, such as acetylcholine or norepinephrine; iii) the brain metabolic phenotype influences the neuroendocrine system via the hypothalamus, modifying the pituitary hormone release. Modified from reference [Citation59] with permission of Springer.
![Figure 4. Brain metabolic phenotypes are reflected in the periphery via three mechanisms: i) individual metabolites distribute to CSF, plasma and urine, and become integrated in the peripheral metabolic phenotype; ii) the brain metabolic phenotype affects the peripheral nervous signaling, thereby controlling the release of peripheral metabolites, such as acetylcholine or norepinephrine; iii) the brain metabolic phenotype influences the neuroendocrine system via the hypothalamus, modifying the pituitary hormone release. Modified from reference [Citation59] with permission of Springer.](/cms/asset/02f425ae-24d4-4819-be56-ed29ac254b97/iedc_a_1446935_f0004_oc.jpg)
