Figures & data
Figure 1. MEDI9447 is a non-competitive inhibitor of CD73 hydrolysis of AMP. (A) The enzyme kinetics of CD73 phosphohydrolysis of AMP were measured in the presence of MEDI9447 or an isotype-matched control mAb using the Malachite Green assay. Increasing AMP substrate concentration does not prevent inhibition of hydrolysis by MEDI9447, indicating the mAb acts as a non-competitive inhibitor. (B) In contrast, the inhibitory activity of APCP can be overcome by increasing the concentration of substrate to outcompete APCP for binding the active site. Error bars represent standard deviation (SD) from triplicate measurements.
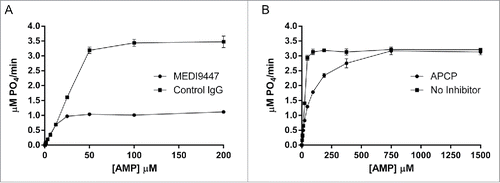
Figure 2. HDX heat map depicting those regions of CD73 (PDB code 4h2f) that undergo differential deuterium uptake when bound to MEDI9447. Relative exchange between antibody-bound and unbound CD73 is depicted as a function of deuterium exposure time with decreased exchange in magenta, increased exchange in blue, and no change in white. The N-terminal region at positions 132–143 exhibits the greatest decrease in exchange at the shortest exchange time point (left image, 30 sec) whereas region 182–187 exhibits the highest degree of differential exchange at the longest time point (right image, 120 min). Regions colored in light brown correspond to residues not detected in the mass spectrometry analysis (2.5% of total sequence). The orientation is such that the N-terminus is at the top and C-terminus is at the bottom of the structure. Color scale bar represents relative % change in deuterium uptake by CD73 in MEDI9447-bound vs. unbound states.
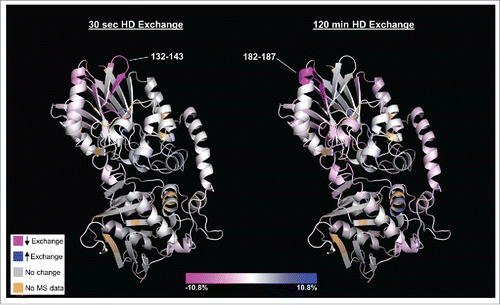
Figure 3. The MEDI9447 epitope resides within the N-terminal domain of CD73. Wild-type (A) and knock-out mutant CD73 proteins (B-F) were immobilized via their HisCitation6 tag on a HTG sensor chip and then binding of MEDI9447 dilutions (5 nM to 0.3 nM, except for (E) at 20 nM to 1.25 nM) was measured by SPR. The mutations V144K (B), K180A (C), N185G (D), and V144K+K180A (E), all reduce MEDI9447 binding. Combining N185G together with either V144K (F) or K180 (data not shown) abolishes binding. (G) SPR kinetics of MEDI9447 binding to wild-type and mutant CD73 proteins. *2:1 fit (see Methods).
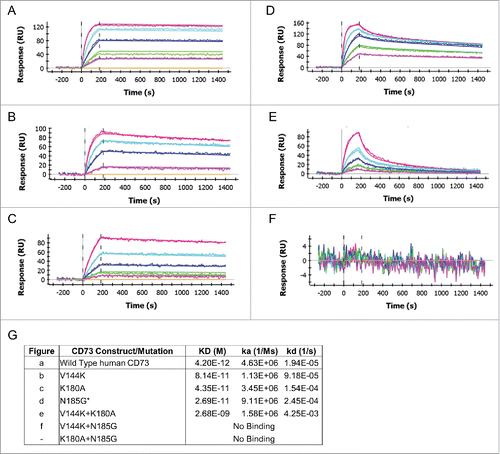
Figure 4. The MEDI9447 epitope is positioned at the apex of the N-terminal domain. (A) Evaluation of MEDI9447 binding to a panel of CD73 knockout and knock-in variants (see Fig. S2 and Supplementary Table 1) revealed 6 residues that constitute the interaction site. Two of the 3 most impactful residues (magenta) are located outside the HDX interface regions (gray). Three less crucial residues (pink) are located within the HDX interface. (AA, amino acid). (B) Knocking in N185 and V144 (K180 is conserved) to a CD73 construct encoding chicken N- and C-terminal domain sequence confers binding to within fold20- the KD for wild-type human CD73 (MEDI9447 dilutions from 5 nM to 0.3 nM; compare to ). (C) Crystal structure of the open conformation of CD73 showing the position of the epitope at the apical, lateral surface of the N-terminal domain. The CD73 N-terminal domain, linker region, and C-terminal domain are depicted in yellow, orange, and blue, respectively. Residues mediating binding are colored as in (A) and shown in blowout, top view. The location of the MEDI9447 epitope is distant from the substrate binding site (adenosine depicted in spheres, C-terminal domain) and the zinc ion coordination site (side chains in cyan, N-terminal domain).
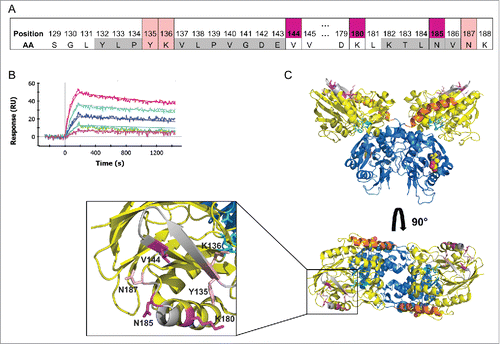
Figure 5. MEDI9447 inhibits the transition of sCD73 to the conformationally active structure. (A) Dose response of MEDI9447 IgG, Fab, or control IgG on the inhibition of sCD73 hydrolysis of AMP as measured using the CellTiterGlo assay. MEDI9447 IgG reaches maximal inhibition at a 1:1 molar stoichiometry with CD73 dimer (arrow). At high concentrations, where MEDI9447 IgG is in excess(>10 nM), a loss of inhibition or “hook effect” is observed. MEDI9447 Fab and control IgG do not inhibit sCD73. Assay was performed using the CellTiterGlo assay as described in Methods (RLU, relative light units). (B) Wild type CD73 was immobilized on a HIS2 biosensor and binding of MEDI9447 (blue sensorgram) and anti-CD73 mAb A (brown sensorgram) was measured by BLI. CD73 pre-incubated with Zn2+ and APCP retains binding to MEDI9447 (black sensorgram) but mAb A binding is lost (orange sensorgram). (C) Although Zn2+ and APCP pre-incubation with CD73 cause a loss in mAb A binding (orange sensorgram), pre-incubation with MEDI9447 before addition of Zn2+ and APCP restores binding (black sensorgram). Binding of mAb A to CD73 alone, CD73 pre-incubated with MEDI9447 (but not Zn2+ and APCP), and MEDI9447 is shown in the brown, blue, and purple sensorgrams, respectively. (D) Cartoon model depicting how MEDI9447 (not depicted) may prevent CD73 from adopting the fully closed, active conformation induced by Zn2+ and APCP; mAb A (red) only binds to CD73 in the open and “intermediate” conformation. Error bars (A), and dots (B,C) represent SD.
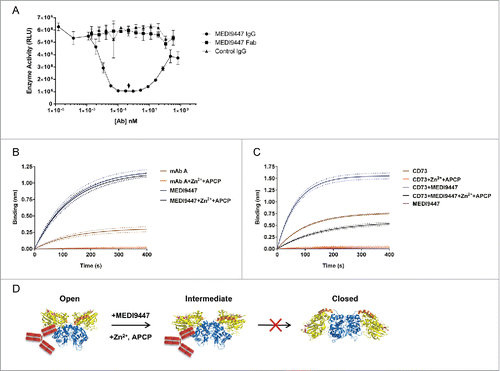
Figure 6. MEDI9447 forms inter-dimer bridges between soluble CD73 molecules. CD73 was incubated with varying amounts of MEDI9447 or anti-CD73 mAb B and analyzed by SEC-MALS. Shown in (A,B) are SEC UV chromatograms with protein retention time on the x-axis and molar mass distributions determined by MALS indicated across the elution profiles with the corresponding mass on the y-axis. (A) At a 1:1 molar ratio (blue trace), MEDI9447 forms complexes with CD73 of ∼1.7 (ˆ) and ∼0.66 (+) megadaltons. Comparably sized complexes are formed at lower ratios of MEDI9447:CD73 (0.5:1 in magenta, 0.1:1 in brown). MEDI9447 and CD73 alone are represented by the black and teal UV traces, respectively. (B) Top-down view of the crystal structure of CD73 dimer showing the mAb B binding region (purple) and MEDI9447 epitope (magenta and pink). (C) CD73 bound to mAb B forms a single predominant complex of ∼270–290 kD (peak at ∼7.2 min). UV traces shown represent 1:1 mAb B:CD73 (blue), 0.5:1 (magenta), and 0.1:1 (brown). mAb A and CD73 alone are in black and teal, respectively.
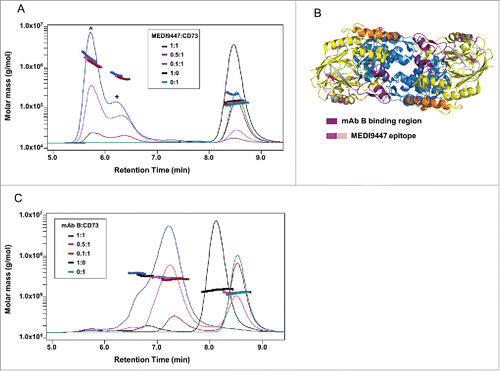
Figure 7. Surface-bound CD73 is inhibited by both IgG and Fab formats of MEDI9447. (A) CD73 was immobilized via a C-terminal histidine tag to a nickel-coated microtiter plate and inhibition of AMP hydrolysis by MEDI9447 IgG, Fab or control antibodies was measured using the Malachite Green assay. MEDI9447 IgG, but not control IgG, inhibits immobilized CD73 hydrolysis of AMP in a dose-dependent manner. MEDI9447 Fab also inhibits CD73 activity, but to a lower extent. (B) When MEDI9447 Fab (cyan) is bound to one arm of an anti-Fd antibody (xFd, gray) and the other arm bound to a non-specific polyclonal Fab (pFab, orange, (B), left cartoon) inhibition is increased to that comparable with MEDI9447 IgG through increasing the effective size rather than valency. (C) Enzyme activity of endogenously expressed CD73 in MDA-MB-231 cells measured by CellTiterGlo assay. As with immobilized recombinant CD73, MEDI9447 IgG inhibits AMP hydrolysis to a greater degree than the Fab, and increasing the effective size of the MEDI9447 Fab by complexing with an anti-Fd antibody enhances inhibition. (D) To test whether the xFd+MEDI9447 can inhibit soluble CD73, AMP hydrolysis was measured using the Malachite Green assay. MEDI9447 Fab either alone or bound to a single xFd arm does not inhibit soluble CD73 activity. In contrast, MEDI9447 Fab binding to both xFd arms (MEDI9447 Fab + xFd, (B), right cartoon) confers bivalency, resulting in CD73 inhibition. Error bars represent SD.
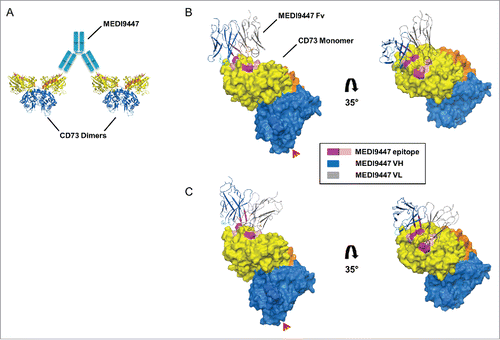
Figure 8. Computationally predicted models of MEDI9447 Fv bound to CD73. (A) Cartoon depiction of how CD73 inter-dimer bridging would be facilitated by the MEDI9447 IgG (cyan) VH and VL interacting with the lateral and medial edges of the binding site, respectively. (C,B) Pose cluster examples showing the Fv model structure extending outwards from the CD73 monomer, though at different projection angles relative to the binding site (CDR H1, H2, H3 in cyan, teal, and blue, and CDR L1, L2, L3 in black, orange, and brown, respectively; Chothia annotation). Color coding of the CD73 structure is as in . The Fv VH and VL are represented in gray and dark blue, respectively. Red arrowheads indicate the GPI anchor attachment site.