Figures & data
Figure 1. Overview of C5 complement activation. Complement C5 activation can occur either by the classical, lectin and alternate complement pathways to generate C5 convertases, or via proteases extrinsic to the complement pathway. C5 convertases or extrinsic proteases can cleave C5 to form proteolytic fragments C5a and C5b. C5a is a potent anaphylatoxin that is rapidly deactivated by removal of the C terminal arginine to form C5adesArg. C5a binds with high affinity (thick black arrows) to both C5aR1 and C5aR2 receptors, while C5adesArg binds with high affinity to the C5aR2 receptor and with low affinity (dashed arrow) to the C5aR1 receptor. The C5aR1 receptor primarily drives pro–inflammatory effects, while the C5aR2 receptor can mediate pro– or anti–inflammatory effects depending on its cellular context. In the absence of cell associated complement inhibitors, C5b interacts with a single copy of complement proteins C6, C7, C8 and a further 18 copies of C9 to form a pore within the cell membrane called the membrane attack complex (MAC).Citation8 The MAC can lyse certain Gram–negative bacteria, promoting bacterial clearance, while host cells can avoid lysis due to the presence of membrane associated complement regulators.
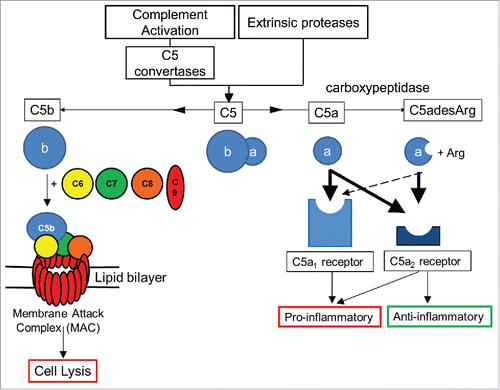
Table 1. Affinities and binding kinetics of MEDI7814. C5a affinity was estimated using surface plasmon resonance (BIAcore). C5 affinity was estimated by kinetic exclusion assay (KinExa). Affinity results are an average of 2 or more separate experiments. N.D. Not determined.
Figure 2. MEDI7814 is specific for human C5a and C5. Representative results showing that MEDI7814 does not bind to related human complement proteins: C3a, C3, C4a and C4. Purified complement proteins were titrated into a DELFIA® biochemical competition assay measuring MEDI7814 IgG binding to 1.2 nM biotinylated human C5a. MEDI7814 binds to human C5a (•) and C5 (▪) as both proteins compete for binding to biotinylated C5a. No competition was observed with complement C3, C3a, C4 and C4a indicating that MEDI7814 does not bind these complement family members when tested at a concentration in excess of 1000-fold over the biotinylated C5a concentration. Data points represent the mean of duplicate wells ± standard deviation. Geomean IC50 values for human C5a and human C5 were 0.3 nM and 0.6 nM respectively (n = 3).
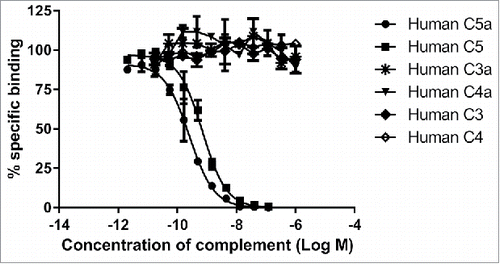
Figure 3. Amino acid sequence alignment of ab116 and MEDI7814 variable domains. The 6 amino acids that differ between ab116 and MEDI7814 variable domains differ are highlighted in red. Complementarity determining regions (CDRs) are defined by black lines and residues are numbered according to Kabat.Citation44
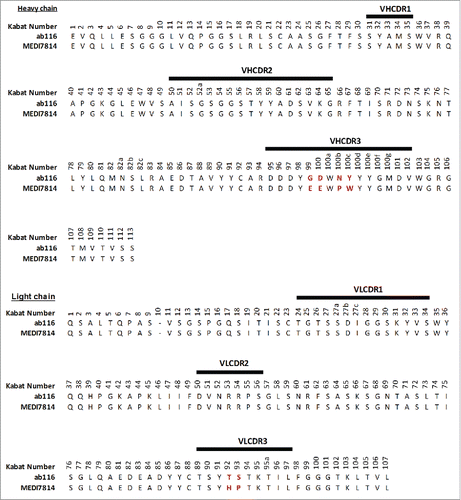
Figure 4. MEDI7814 inhibits fluorescently labelled C5a binding to C5aR1 and C5aR2 receptors. Representative results showing that MEDI7814 (▴) inhibits (A) Alexa® Fluor 647 labelled human C5a (AF647 C5a) binding to human C5aR1 receptor, (B) AF647 human C5a binding to human C5aR2 receptor and (C) AF647 human C5adesArg binding to human C5aR2 receptor. The isotype matched irrelevant control (▪) did not inhibit C5a ligand binding. C5a receptors were expressed in stably transfected HEK293 cells and C5a ligand binding was measured using Fluorimetric Microvolume Assay Technology (FMAT). Data points represent the mean of duplicate wells ± standard deviation. IC50 values were not determined as the affinity of MEDI7814 exceeds the concentrations of C5a/C5adesArg in the assays. The effect of MEDI7814 on the binding of C5adesArg to C5aR1 receptor was not determined due to the low affinity of C5adesArg for this receptor.
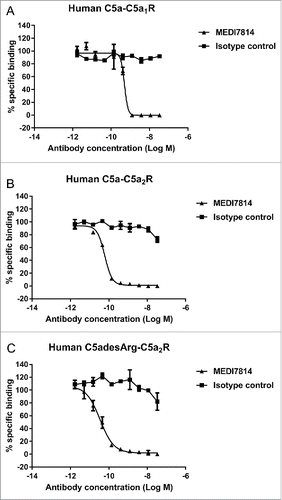
Figure 5. MEDI7814 neutralizes C5a and C5adesArg responses in vitro. Representative results showing that MEDI7814 (▴) neutralizes calcium mobilisation in HEK 293 expressing human C5aR1 receptor and Gα16 using (A) 100 pM human C5a, (B) 400 pM human C5adesArg. Data points represent the mean of duplicate wells ± standard deviation. (C) MEDI7814 neutralizes 2 nM human C5a induced chemotaxis of human neutrophils. Data points represent the mean of duplicate wells ± standard deviation. MEDI7814 IC50 values were not determined for A, B and C as the affinity of MEDI7814 exceeds the concentrations of C5a/C5adesArg in these assays, n = 2. No inhibition of responses were observed with the isotype matched irrelevant control antibody (▪).
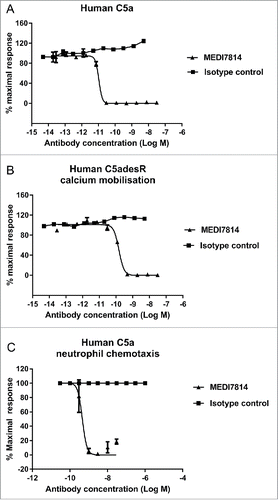
Figure 6. MEDI7814 neutralizes C5a-C5aR1 receptor mediated CD11b upregulation in human whole blood. Representative results showing that (A) MEDI7814 (▴) neutralizes endogenous C5a mediated neutrophil CD11b up-regulation in human whole blood with a geomean IC50 of 28 nM. Endogenous C5a was generated by activation of complement using E coli as described by Fung et al.Citation45 Data points represent the mean ± standard error of the mean for 5 separate experiments. No inhibition of response was observed with the isotype irrelevant control antibody (▪). (B) CD11b upregulation is mediated predominately via the C5aR1 receptor. 100 nM of an anti-C5aR1 receptor neutralizing antibody (anti-C5aR1 F(ab')2) inhibits C5a mediated CD11b up-regulation in whole human blood upon complement activation using E.coli. 100 nM of an anti-C5aR2 receptor neutralizing antibody (anti-C5aR2 F(ab')2) and an isotype negative control F(ab')2 showed no significant inhibition, indicating that CD11b up-regulation is mediated predominately through the C5aR1 receptor. 100 nM MEDI7814 inhibits CD11b up-regulation, whereas the isotype control antibody showed no inhibition. T0 and T10 represent CD11b level at time 0 and 10 minutes after blood was incubated at 37°C without E.coli. Individual data points from ≥ 6 different donors are shown alongside the mean ± standard deviation for each test condition. ****p < 0.0001, by one-way ANOVA followed by Sidak's multiple comparisons test, (ns, not significant).
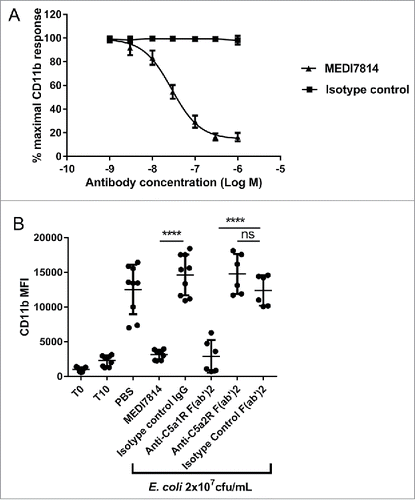
Figure 7. MEDI7814 neutralizes C5a mediated HMGB1 release in human whole blood. Representative results showing that 100 nM MEDI7814 inhibits HMGB1 release from human whole blood upon complement activation using E.coli. Individual data points from ≥ 4 different donors are shown alongside the mean ± standard deviation for each test condition. *p < 0.05, by Kruskal-Wallis test followed by Dunn's multiple comparisons test, (ns, not significant). The Kruskal-Wallis non-parametric test was used as the number of samples tested was low for some groups and therefore no assumptions could be made about the normal distribution. The “-E.coli/-Ab” control contained PBS in place of E coli. and test antibody, and the “+E.coli/-Ab” control contained PBS in place of test antibody.
Figure 8. MEDI7814 does not inhibit bacterial cell killing and clearance. Representative results showing that MEDI7814 (▴) does not inhibit (A) lysis of luminescent K. pneumoniae, in the presence of human serum. No effect was observed with the isotype irrelevant control mAb (▪), whereas the positive control mAb, Eculizumab (•) effectively inhibited cell lysis. Data points represent the mean of duplicate wells ± standard deviation. (B) MEDI7814 does not inhibit C3b mediated opsonophagocytic killing (OPK) of a luminescent strain of P. aeruginosa in human whole blood. Blood was treated with 600 nM MEDI7814 or an IgG4 isotype negative control antibody prior to being added to wells containing a positive control anti-CD11b mAb or an IgG1 isotype irrelevant control mAb and P. aeruginosa. As MEDI7814 shows a mean luminescence similar to the negative PBS control (4593 RLU) in the presence of the IgG1 irrelevant control mAb, this indicates that it does not inhibit OPK. The anti-CD11b mAb blocks OPK resulting in an increased luminescence. Data points represent mean RLU ± standard error of the mean for 4 different donors.
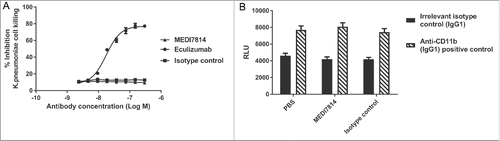
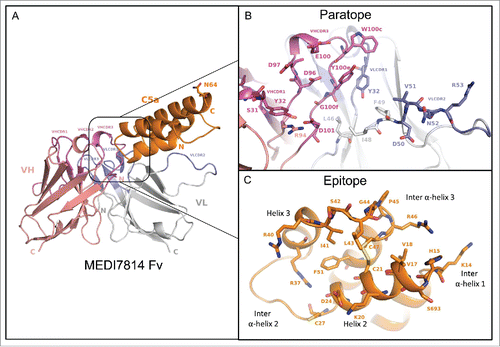
Figure 9. Structure of the human C5a/MEDI7814 scFv complex. (A) Left: Overall complex structure showing C5a (orange) interacts primarily through the VHCDR3 and VLCDR2 of MEDI7814 scFv: variable heavy chain domain (salmon pink), VHCDRs (magenta), variable light chain domain (grey) and VLCDRs (blue), C5a glycosylation site N64 is shown as a stick. (B) Paratope residues (Kabat) of MEDI7814 scFv: VHCDR1 (S31, Y32), VHCDR3 (D96, D97, E100, W100c, Y100d, Y100e, G100f, M100g, D101), VLCDR1 (K31, Y32), VLCDR2 (D50, V51, N52, R53), VH framework 3 (R94), and VL framework 2 (L46, I48, F49). The framework amino acids are Vernier residues, important for the structural integrity of antibodies.Citation50 (C) Discontinuous epitope on C5a recognized by MEDI7814 scFv comprises amino acid residues Y13-C21, D24 and G25, C27, R37, R40-C47 and F51, shown as orange sticks.