Figures & data
Figure 1. Relative stability of IgGs toward reduction by the thioredoxin system. IgG1, IgG2, and IgG4 mAbs were incubated with the components of the thioredoxin system. The figure contains data for 4 distinct IgG1s, 3 distinct IgG4s, and 2 distinct IgG2s. Samples were taken at the indicated timepoints and intact mAb was quantified using capillary electrophoresis. Experiment was repeated twice with single replicates; 1 replicate is shown.
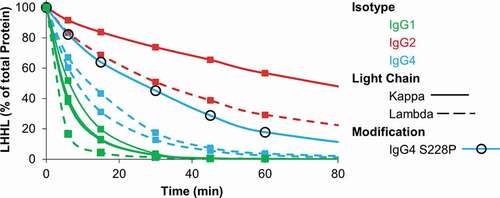
Figure 2. Intermediate species formed during reduction by the thioredoxin system. IgG1, IgG2, and IgG4 mAbs were incubated with the components of the thioredoxin system. Samples were taken at the indicated timepoints and mAb fragments were quantified using capillary electrophoresis. For all figures intact mAb (LHHL) is on the left axis and fragment species (HHL, HH, and HL) are on the right axis. Free heavy and light chain were not shown to aid in visualization, they are however, included in Fig S1. Experiment was repeated twice with single replicates; 1 replicate is shown.
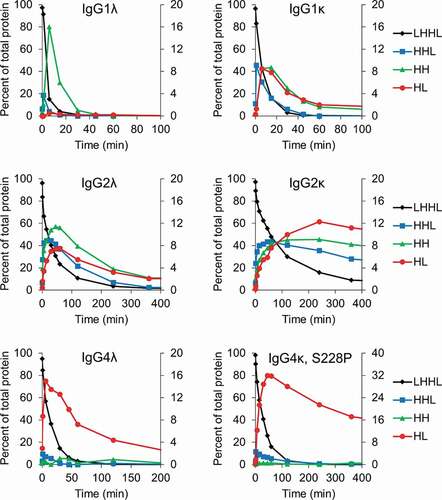
Figure 3. Cartoon depiction of the major intermediates formed for different IgG isotypes when exposed to reducing conditions.
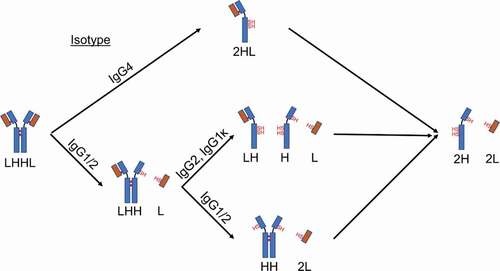
Table 1. Amino acid sequence comparison of the hinge region for IgG1, IgG2, and IgG4 antibodies using the EU index for antibody numbering. The positions of the mutations evaluated are indicated in red.
Table 2. The relative stability of the IgG4λs was assessed by fitting the results in to a first-order exponential decay to determine the half-life of each mAb. An IgG1λ and IgG2λ were included for comparison
Figure 4. Hinge mutations increase stability of IgG4 mAbs toward reduction. IgG4 mAbs with the indicated hinge mutations were incubated with the components of the thioredoxin system. Samples were taken at the indicated timepoints and intact mAb was quantified using capillary electrophoresis. Data represent the mean ± SD of triplicate experiments.
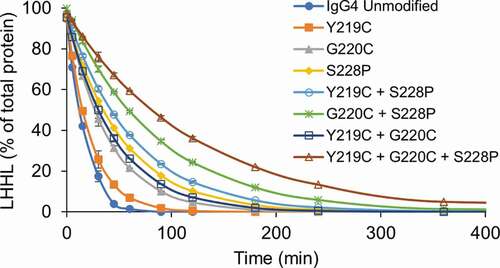
Figure 5. Hinge mutations decrease half-antibody formation. IgG4 mAbs with the indicated hinge mutation(s) were incubated with the components of the thioredoxin system. Samples were taken at the indicated timepoints and mAb fragments were quantified using capillary electrophoresis. Free heavy and light chain were not shown to aid in visualization, they are however, included in Fig S3. Data represent the mean ± SD of triplicate experiments.
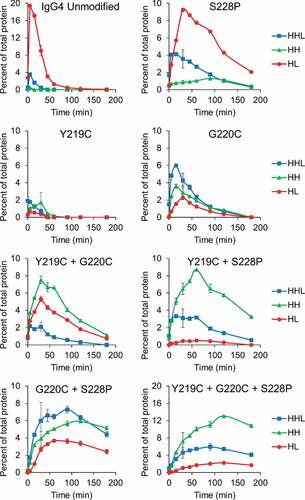
Figure 6. Hinge mutations decrease Fab-arm exchange. (a) Quantification of hybrid mAb formation for IgG4 mAbs with the indicated hinge mutation(s) after incubation with a second, unmodified IgG4. Hybrid mAb was quantified using capillary electrophoresis and quantification of each parent mAb is included in Fig. S4. Data represent the mean ± SD of triplicate experiments. (b) Example electropherogram from the capillary electrophoresis for the unmodified IgG4. (c) Example mass spectrometry data for the unmodified IgG4. Mass spectrometry for remaining samples are in Fig. S5 and S6.
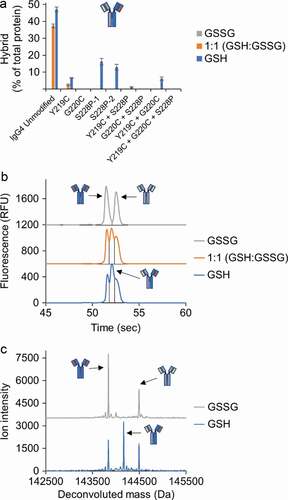
Figure 7. Hinge mutations do not affect antigen, FcRn or FcγRIIIa binding in vitro. (a) Antigen binding of the IgG4 mAbs with the indicated hinge mutations were compared to the unmodified IgG4 in an ELISA assay. An isotype control demonstrated no binding up to 5000 nM (not shown). Data represent the mean ± SD of duplicate experiments. (b) FcRn and (c) FcγRIIIa binding for the IgG4 s with hinge mutations were compared to the unmodified IgG4 in AlphaLISA assays. Rituximab was run as a known positive control in the FcγRIIIa AlphaLISA. Data represent the mean ± SD of duplicate independent experiments.
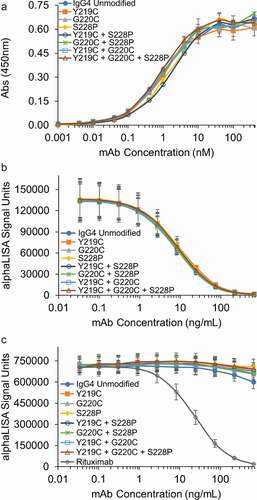
Table 3. Summary of the effect of each mutation on the relative stability toward reduction, half-mAb formation, and Fab-arm exchange
