Figures & data
Figure 1. Overview of the library construction process.
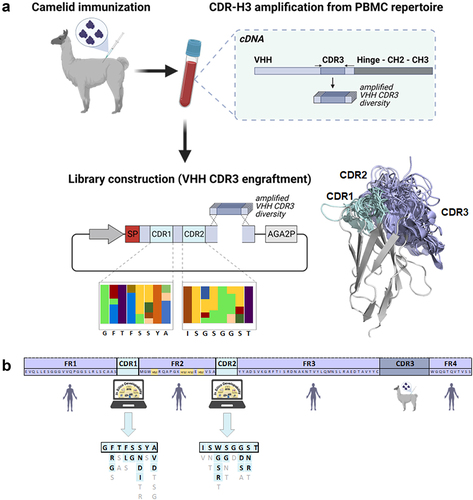
Figure 2. Amino acid distribution of CDR1 and CDR2 of non-immunized llamas compared with artificially designed diversities and observed compositions in humanized sdAb libraries.
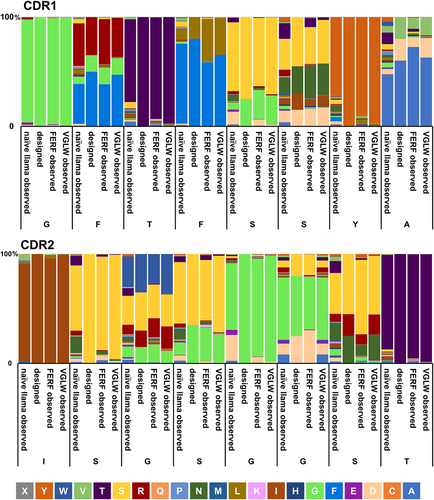
Figure 3. Yeast surface display enables the isolation of (rh) NKp46-targeting sdAbs from both humanized libraries.
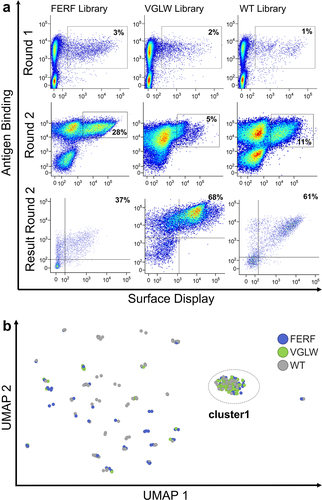
Table 1. CDR1–3 and hallmark (HM) sequences of immunized llama WT or humanized VHH libraries (FERF or VGLW) that were selected for production and experimental profiling (shown in ). Residues that are different from the most potent sequences within each CDR3 sequence cluster are shown in orange; susceptible chemical liability and post-translational modification motifs (DG, NG, M, unpaired C, and NXS/T) are indicated as red boxes.
Table 2. In silico developability assessment of VHHs obtained from different library approaches. sdAbs were analyzed for their sequence identity compared to the most similar human germline (MOST SIMILAR GERMLINE) either based on the entire variable chain region (SEQ-ID) or the framework region only (SEQ-ID FR), as well as for their total number of specific chemical liabilities and PTMs, i.e., non-canonical cysteines, methionine oxidations, asparagine deamidations or aspartate isomerizations, and N-glycosylations, in structurally exposed CDR residues as derived from automatically generated models. As calculated physicochemical developability descriptors (IN SILICO PHYSCHEM), structure-based pI values (pIfv 3D), the AggScores of the entire variable regions, and the AggScores of CDR regions only (CDR AggScore), as well as the positive patch energy of the CDRs (CDR positive patch energy), are shown. The complementing color coding indicates scores within one standard deviation from a benchmark mean (dataset of 77 biotherapeutics approved for human application) as green, scores above one standard deviation as yellow, and scores above two standard deviations as red. For the AggScores, this classification was slightly adjusted based on correlation analyses to internal experimental HIC data.
Table 3. Analytical and early developability data of one-armed constructs organized by cluster and affinity, including SEC purity, HIC retention time, mean Tonset, and dissociation constant measured via BLI.
Figure 4. Binding capacities of humanized sdAbs and WT VHHs reformatted as one-armed SEEDbodies as determined by BLI.
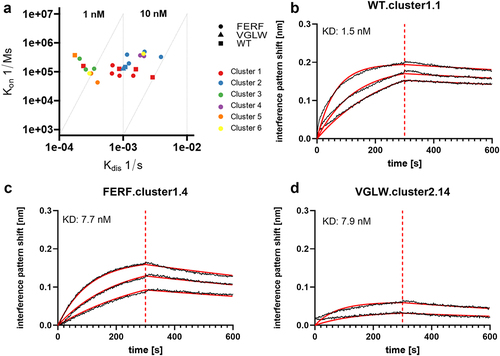
Table 4. Production yields and analytical purity of nominated sequences and their ability to function as NKCE with humanized cetuximab (ctx) on the GA chain, as indicated by the EC50 for killing of target cells. Evaluation of AC-SINC is included for the one-armed constructs.
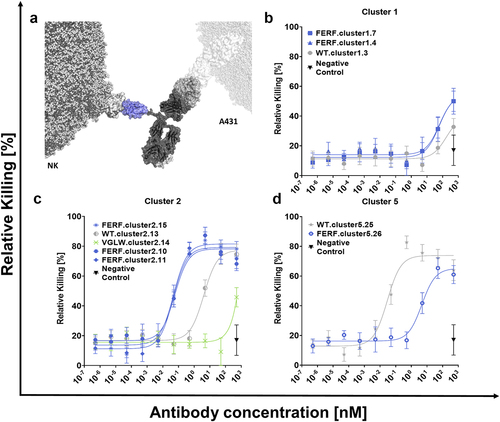
Figure 5. NK cell engagers (NKCEs) harboring de novo humanized sdAbs from the FERF design trigger significant NK cell mediated killing of EGFR overexpressing tumor cells.