Figures & data
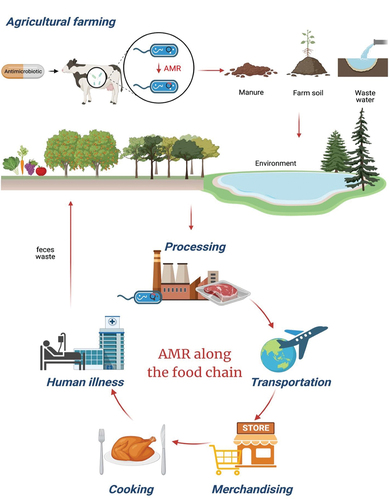
Figure 2. The mechanism of antibiotic action (left). Antibiotics can inhibit the growth of bacteria by interfering with (1) the bacterial cell wall or (2) the cell membrane synthesis. Antibiotics also target (3) protein synthesis and (4) nucleic acid. (5) lastly, antibiotics can also function as antimetabolites by inhibiting the folate metabolism via a pathway involving para-aminobenzoic acid (PABA) and two folic acid precursors, dihydrofolate (DHF) and tetrahydrofolate (THF). Sulfonamide and trimethoprim are structural analogs of PABA and DHF that block dihydropteroate synthetase and dihydrofolate reductase, respectively, preventing bacteria from synthesizing folic acid, a component required for the synthesis of nucleotides. On the other hand, there are five categories of AMR mechanisms (right). (1) the efflux pumps export antibiotics from the cells to lower the concentration of antibiotics inside bacterial cells. (2) the permeability of the outer membrane can be reduced by porins that allow antibiotics to enter the bacterial cell passively. (3) target modification includes changing the antibiotic targets to lessen antibiotic binding. (4) antibiotics are rendered inactive by enzymes that either break down or alter the antibiotic molecule. (5) target bypass replaces the antibiotics target with a replacement protein that performs the same function without being inhibited by the antibiotics, rendering the antibiotics useless and the original target unnecessary.
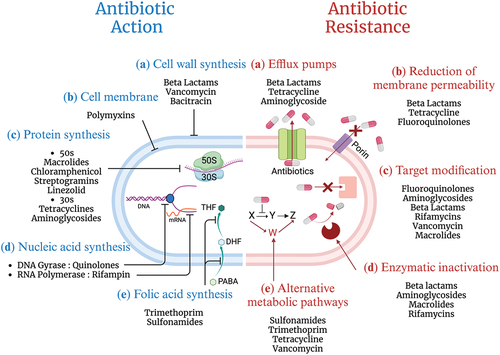
Figure 3. Overview of ML for AMR prediction (a) genome assembly is the process of rebuilding the whole DNA sequence from the short, fragmented sequences. Genome annotation is the act of identifying and elucidating the functionalities of the bacteria. On the other hand, the MALDI-TOF MS, Raman spectroscopy, and FTIR are utilized for preprocessing in the context of spectroscopy-based ML applications. (b) Genome encoding refers to the process of delivering genetic data uniformly so that computers can understand and evaluate it. (c) ML models find underlying patterns of datasets for predicting new, unseen data. (d) Both holdout validation and k-fold cross-validation are crucial strategies for figuring out how well a ML model will perform on new, unseen data.
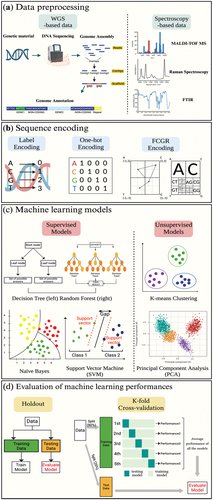
Figure 4. Information available for genomes with the Genome Annotation Service in BV-BRC. (a) The genome landing page. (b) Genome browser. (c) Specialty gene page. (d) Phylogeny.. (e) Circular view of the genome. Researchers can search for genes in their genome that are related to AMR, virulence factor.
Figure 5. The bacterial and viral bioinformatics resource center (BV-BRC) (https://www.bv-brc.org/). (a) BV-BRC’s genome assembly landing page where researchers can assemble either single- or paired-end short reads, or longer reads. On the right side with an enlarged figure, there are listings of the assembly tools available. (b) The genome annotation landing page where researchers can annotate assemblies.
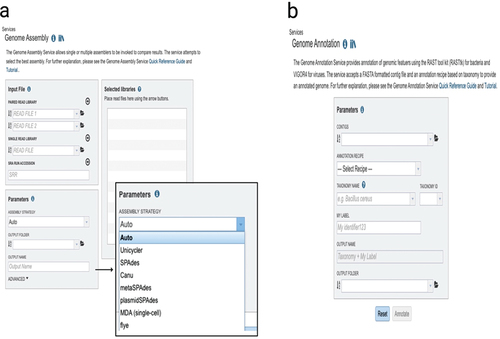
Table.1. Applications of machine learning (ML) using whole genome sequencing (WGS)-based data for AMR prediction.