Figures & data
Figure 1. Western blot confirming binding of mAbs to CPS. B. pseudomallei (Bp) 1026b, B. mallei (Bm) China 7, CPS purified from Bp RR2683, or B. thailandensis (Bt) E264 were incubated with proteinase-K, separated by SDS PAGE, and subsequently transferred to a nitrocellulose membrane. Membranes were probed with 0.5 µg of mAb and binding was detected with an HRP-conjugated goat anti-mouse kappa chain antibody. IgG3 mAb 3C5 and IgG1 mAb 2A5 bind to a high molecular weight, proteinase-K resistant antigen found in B. pseudomallei, B. mallei, and purified CPS. No mAb binding was visualized against B. thailandensis, which lacks CPS.
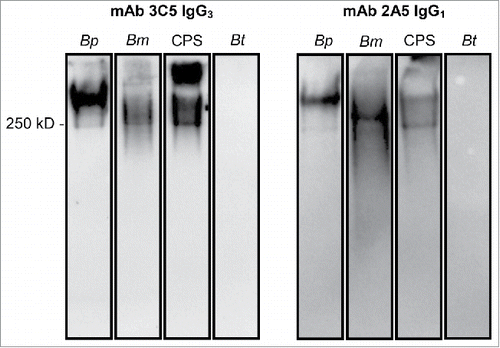
Figure 2. Binding specificity of subclass-switch family mAbs and 3C5 IgG3 F(ab')2 fragments given as a function of response units (RU) generated over time. Data are shown from a representative experiment. A BIAcore X100 instrument was used to determine the affinity of each mAb for CPS. CPS purified from Bp RR2683 was conjugated to biotin and immobilized on a SA sensor chip. Binding was analyzed by injecting 8 samples diluted 2-fold (dilutions; 0.33–333 or 12–3,333 nM) for 60 s, followed by 120 s of passive dissociation.
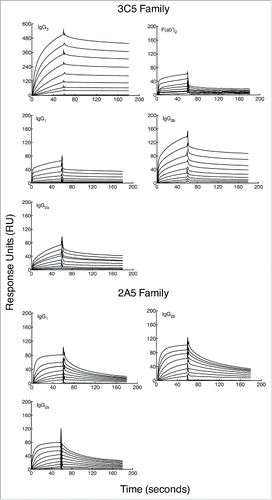
Figure 3. Binding affinity of subclass-switch family mAbs and 3C5 IgG3 F(ab')2 fragments given as a function of RUs generated by concentration. Data are shown from a representative experiment. The steady-state model from BIAevaluation software was applied to each graph in to determine the dissociation constant (KD) of each mAb. A smaller KD corresponds to a higher affinity. Note the higher mAb concentrations needed for calculation of affinity for the 3C5 non-IgG3 mAbs.
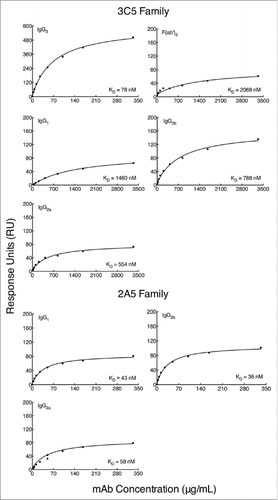
Table 1. Summary of antibodies generated for this study.
Figure 4. Comparison of binding activity within each mAb subclass by Western blot. B. pseudomallei Bp82 total cell lysate (87 μg) was separated by SDS-PAGE and transferred onto nitrocellulose membrane. The membranes were probed with mAb 3C5 family (panel A) and 2A5 family (panel B) using a miniblotter. 3C5 IgG3 shows substantially higher reactivity to CPS compared to the other subclasses. The reactivity between different subclasses of mAb 2A5 and CPS was comparable. 2A5 and 3C5 antibody concentration was prepared from 1mg/ml stock. Exposure time for Western blots in panel A and B were different.
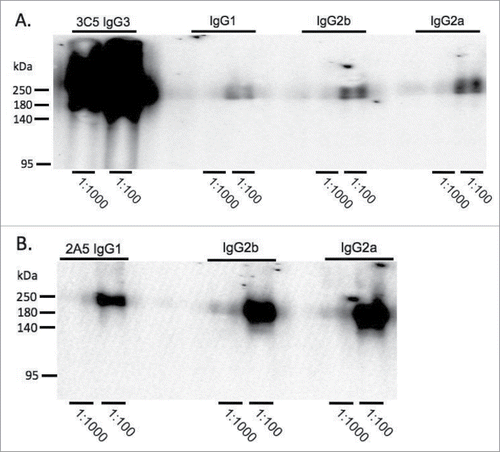
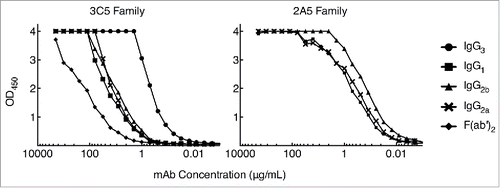
Figure 5. Direct antigen binding ELISA comparing 3C5 IgG3 and 2A5 IgG1 binding with subclass with switch family mAbs. CPS purified from Bp RR2683 was immobilized in the solid phase at 4 µg/mL. Two-fold serial dilutions of mAb or F(ab')2 were added in the fluid phase starting at 2,000 µg/mL and mAb binding was detected with an HRP-conjugated goat anti-mouse kappa chain antibody. All trials were completed in duplicate.