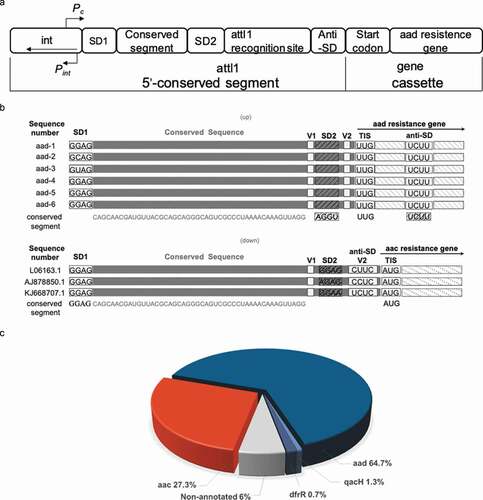
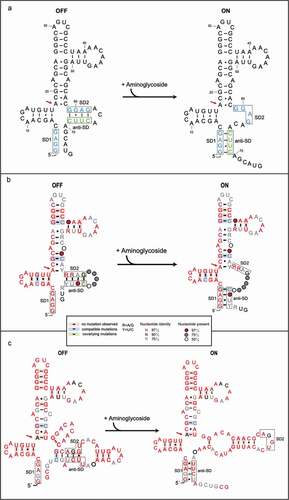
(a) The secondary-structure of the
aac riboswitch from
P. fluorescens in the paper published by Jia et al [
Citation35]. The red arrow indicates the position of the inactive A18G point mutation.
(b) The covariance model of
aac riboswitch in the paper published by Wang et al
(36). Red, black and gray nucleotides indicate the conservation of at least 97%, 90%, and 75%, respectively. The circles with red, gray, white color represent the positions in which nucleotide identity is less conserved as 97%, 75% and 50%, respectively. Base pairs shaded in red exhibit natural covariation, while those in blue exhibit compatible mutations. Purines and pyrimidines are identified by R and Y. The red arrow indicates the position of the inactive A18G point mutation.
(c) The covariance model of the
aad riboswitch RNA. Red, black and gray nucleotides indicate the conservation of at least 97%, 90%, and 75%, respectively. The circles with red, gray and white color represent the positions in which nucleotide identity is less conserved as 97%, 75% and 50%, respectively. Base pairs shaded in red exhibit natural covariation, while those in blue exhibit compatible mutations. Purines and pyrimidines are identified by R and Y. The red arrow indicates the position of the inactive A18G point mutation. The covariant structures were drawn using R2R and Adobe Illustrator.