Figures & data
Table 1. “Core” effectors in different groups of plant pathogens
Figure 1. A schematic representation of in silico prediction and validation of putative CREs in oomycetes. The secretome prediction pipeline begins with the removal of proteins without a signal peptide (SP) while retaining those with a transmembrane domain (TM), by use of signalP tool and THMM, respectively. Effector proteins with a TM are discarded after signal peptide cleavage as these proteins are not likely to be retained in the plasma membrane. This is followed by removing effector proteins without the signature RxLR motif using HMMscan tool. Orthology analysis is performed to determine RxLR effectors that are conserved within strains or within species of a pathogen (CREs) using orthology analyses tools like COG, eggNOG or orthofinder. The final output is composed of putative secreted CREs with a SP, RxLR motif and without a TM. This output is further authenticated through in planta expression to ascertain their role in virulence for instance, their role in enhancing/suppressing host immunity, localization in planta using confocal microscopy as well as interacting proteins within host partners
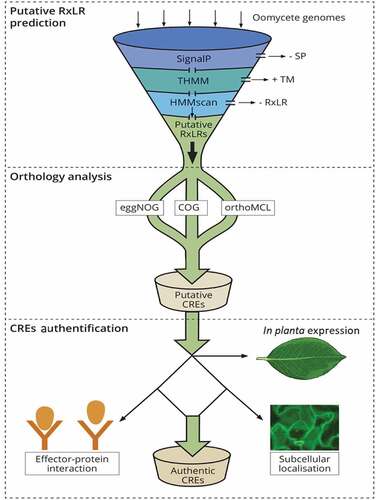
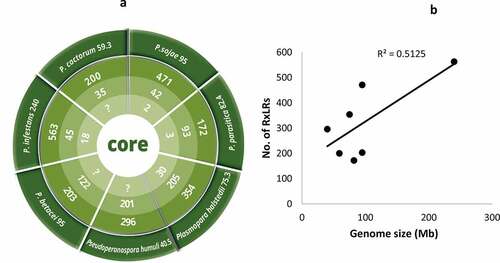
Figure 2. Illustration of phytopathogenic oomycetes with their respective genome sizes (Mb) on the outermost ring. Most of the genomes are Phytophthora spp (p). In terms of genome size, P. infestans and Pseudoperonospora humulis recorded the highest (240Mb) and lowest (40.5Mb) genome sizes, respectively. Counting from the outside, the second ring is the total number of predicted RxLR effectors ranging from 172 in P. parasitica to 563 in P. infestans. The third ring is the total number of putative CREs while the fourth ring is the total number of authentic CREs with Plasmopara halstedii recording a total of 30 CREs. In terms of association between genome size and the number of predicted RxLR effectors in oomycetes, insignificant positive correlation (P = 0,07;R2 = 0.51, at 95% confidence level) was recorded (b)
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and freely available, under a license allowing re-use by any third party for any lawful purpose. Data shall be findable and fully accessible.
