Figures & data
Figure 1. Hypoxia induces the outbreak of ISKNV disease. (A) Infected fish were divided into hypoxia (2 mg/l DO) and normoxia (7 mg/l DO) groups with 30 fish samples for each group. Both groups were cultivated in the experimental tank system setup. as controls, the uninfected fish were cultivated in the same experimental setup. This experimental was repeated thrice. (B) Red line represents infected fish cultured under hypoxia, and blue line denotes infected fish cultured under normoxia. Green line indicates uninfected fish cultured under hypoxic conditions, and black line signifies uninfected fish cultured under normoxia. (C and D) Spleen samples were separated from the mandarin fish at days 0, 3, and 6 p.I. Levels of viral genomic DNA were detected via qPCR, and those of isknv-mcp mRNA were detected via qRT-PCR. (E) Spleen samples were separated from the mandarin fish at 6th day p.I. and then used for biopsy and HE staining.
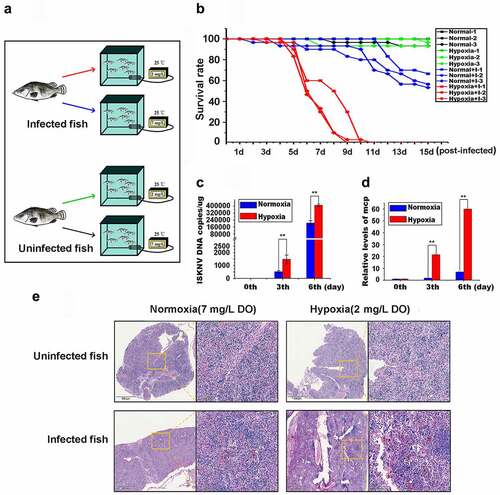
Figure 2. Hypoxia promotes ISKNV replication. (A) Infected cells cultured under hypoxia (3% O2) or normoxia (21% O2). at 0, 12, 24, and 48 h p.i., the total DNA, RNA, protein, and cell lysates were acquired repeated freezing and thawing for three times and isolated for further analyses. (B) Levels of viral genomic DNA were detected via qPCR. (C) Virus titers (TCID50) of the cell lysates acquired by repeated freezing and thawing for three times. (D) Levels of isknv-mcp mRNA were detected via qRT-PCR. (E) Levels of ISKNV VP101R were detected via Western blot.
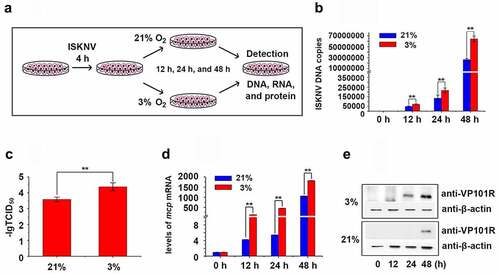
Figure 3. Active HIF pathway promotes ISKNV replication. (A) Infected cells cultured normoxic (21% O2) were treated with 1 mM DMOG. at 0, 12, 24, and 48 h p.i., total DNA, RNA, protein, and cell lysates were acquired via repeated freezing and thawing for three times and isolated for further analyses. (B) Levels of viral genomic DNA were detected via qPCR. (C) Virus titers (TCID50) of the cell lysates acquired by repeated freezing and thawing. (D) Levels of isknv-mcp mRNA were detected via qRT-PCR. (E) Levels of ISKNV VP101R were detected via Western blot.
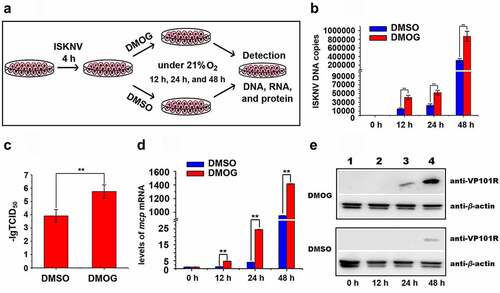
Figure 4. ISKNV HRE-regulated genes respond to the HIF pathway. (A) Cells were co-transfected with 15 predicted pGL3-HREs-luc and pRT-TK plasmids. at 24-h post transfection, dual reporter assay screened the effects of DMOG on all 15 predicted ISKNV HREs in MFF-1 cells. the DMSO-treated cells were used as control. the y-axis represents the luciferase treated withDMOG/luciferase treated with DMSO. (B) MFF-1 cells infected with ISKNV. the cells cultured under normoxic (21% O2) condition were treated with 1 mM DMOG. the DMSO-treated cells were used as control. at 48 h p.i., total RNAs were acquired for further analyses. Levels of isknv orf012r, orf014r, orf019r, orf033r, orf039r, orf063l, orf077r, orf084l, orf085r, orf089r, orf090l, orf097r, orf101l, orf117r, and orf119l. mRNA was detected via qRT-PCR. the y-axis represents the relative expression levels of isknv genes after DMOG treatment/relative expression levels of isknv genes after DMSO treatment.
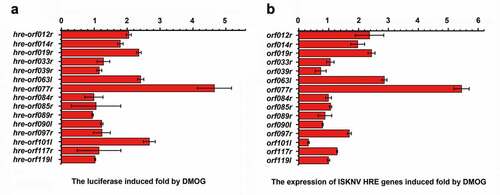
Table 1. Predicted ISKNV-HREs in the ISKNV genome (NC_003494.1)
Figure 5. ISKNV HRE-regulated genes respond to HIF pathway. (A) Potential HIF binding sites in the promoter regions of ISKNV orf077r. Two putative HIF binding site motifs were identified (HRE-1 and HRE-2). the sequences of a double-stranded oligonucleotide probe containing the HRE-1 and HRE-2 were used for EMSA. (B) MFF-1 cells were transfected with pGL3-orf077r HREs-luc, pGL3-orf077r HREs-ΔHRE-1 or pGL3-orf077r HREs-ΔHRE-2, and the pRT-TK plasmids were co-transfected as inner control. Effects of DMOG on the promoter activities of full length orf077r HREs, orf077r HREs-ΔHRE-1, and orf077r HREs-ΔHRE-2 in MFF-1 cells as detected by dual reporter assay. (C) EMSA assay confirmed that the HIF-1α protein binds to HRE-1 and HRE-2. Cells expressing HIF-1α-GFP. at 24 h post transfection, DMOG was used to activate the HIF-1 pathway, and nuclear proteins were extracted using EMSA assay after 12 h of DMOG treatment. (D) Active HIF pathway induced the expression of orf077r in vitro. Infected cells cultured normoxic (21% O2) condition were treated with 1 mM DMOG. at 0, 12, 24, and 48 h p.I., total RNAs were acquired via repeated freezing and thawing for three times and isolated for further analyses. Levels of orf077r mRNA were detected via qRT-PCR. (E) Hypoxia induces the expression of orf077r in vitro. Cells were infected with ISKNV and then cultured under hypoxia (3% O2) or normoxia (21% O2). at 0, 12, 24, and 48 h p.i., total RNAs were acquired via repeated freezing and thawing for three times and isolated for further analyses. Levels of orf077r mRNA were detected via qRT-PCR. (F) Hypoxia induces the expression of orf077r in vivo. Mandarin fish were infected with ISKNV and then cultured under hypoxia (2 mg/l DO) and normoxia (7 mg/l DO). at 0, 3, and 6 days p.i., total RNAs were acquired via repeated freezing and thawing for three times and isolated for further analyses. Levels of orf077r mRNA were detected via qRT-PCR. (G-H) Cells expressing VP077R-GFP were infected with ISKNV. at 48 h p.i., total DNA and RNA were acquired. Levels of viral genomic DNA were detected via absolute qPCR. Levels of isknv-mcp mRNA were detected via qRT-PCR.
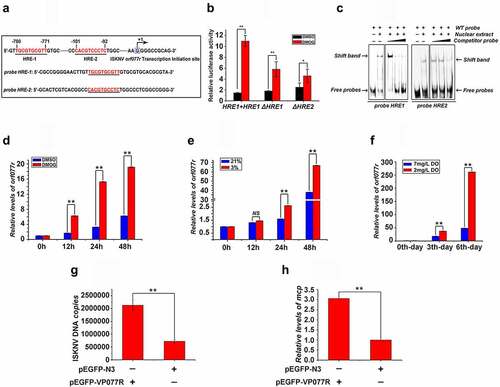
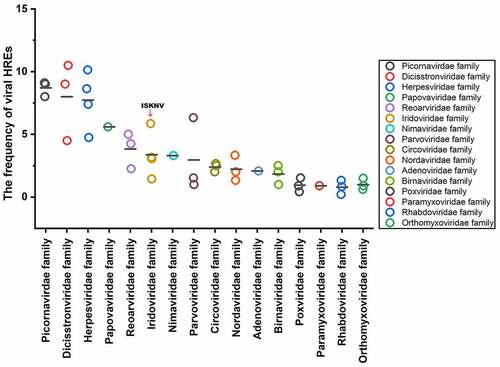
Figure 6. Frequency of HREs in aquatic animal viruses from 16 families. HRE frequency of each virus: carp picornavirus 1 (9.0), clownfish picornavirus (9.0), bluegill picornavirus (8.0), mud crab dicistrovirus (4.5), Macrobrachium rosenbergii Taihu virus (9.0), taura syndrome virus (10.5), abalone herpesvirus (10.14), cyprinid herpesvirus 3 (8.63), anguillid herpesvirus 1 (7.4), ictalurid herpesvirus 2 (4.74), marbled eel polyomavirus (5.6), grass carp reovirus (5.0), Scylla serrata reovirus (2.25), piscine reovirus (4.25), infectious spleen and kidney necrosis virus (5.86), Singapore grouper iridovirus (3.05), lymphocystis disease virus 1 (1.44), frog virus 3 (3.16), white spot syndrome virus (3.3), tilapia parvovirus (6.33), infectious hypodermal and hematopoietic necrosis virus (1.0), sea star-associated densovirus (1.53), clinch densovirus 1 (2.8), uncultured virus clone AfaCV3 (2), uncultured virus clone SdaCV2 (2.5), Anguilla anguilla circovirus (2.66), red-spotted grouper nervous necrosis virus (3.33); tiger puffer nervous necrosis virus (2.0); covert mortality nodavirus (1.33), white sturgeon adenovirus 1 (2.08), spring viremia of carp virus (.2), viral hemorrhagic septicemia virus (.83), hirame rhabdovirus (1.33), infectious salmon anemia virus (.2), Tilapia lake virus (.9), pilchard orthomyxovirus (1.5), carp edema virus (.41), salmon gill poxvirus (1.53), cheloniid poxvirus 1 (.9), Atlantic salmon paramyxovirus (.89), infectious pancreatic necrosis virus (1), Tasmanian aquabirnavirus (2), and Tellina virus 1 (2.5).
Supplemental Material
Download Zip (583.6 KB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.