Figures & data
Figure 1. The capsule and pathogenicity of strain WUSS351. (a) The phylogenetic tree of S. suis cps clusters from different serotypes or genotypes, created using the Neighbor-Joining method. (b) The comparative analysis of cps gene clusters between S. suis NCL4 strains WUSS351 and YS73. (c) The capsule of S. suis strains WUSS351 and SC070731 (an encapsulated strain control), as observed using TEM. (d) The survival curve of mice injected with strains WUSS351 and SC070731 (a virulent strain control) at the dose of 3 × 108 CFU/mouse or PBS (a negative control). Asterisks indicate pairs of significantly different values (n = 10, the Log-rank (Mantel-Cox) test, **** indicates p < 0.0001, ns indicates not significant).
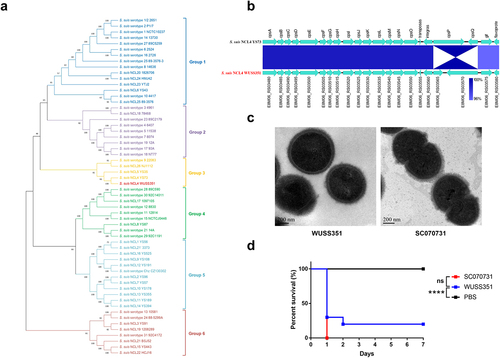
Table 1. MICs of strain WUSS351
Figure 2. Comparative genome analysis. (a) The genetic distance among S. suis strains was calculated using the OrthoANIu algorithm. Based on the distance data, Neighbor-Joining method was used to construct the phylogenetic tree. S. pneumoniae strain TIGR4 was used as an out-group. (b) The Mauve software was used to align 52 S. suis genomes, and the Circos software was used to display comparison results. To better present the comparative genome analysis results, strain WUSS351 and representative strains from different branches of the phylogenetic tree shown in Figure 2a were selected. A 25 kb genomic island was predicted in the genome of strain WUSS351 but not in the other 51 S. suis strains.
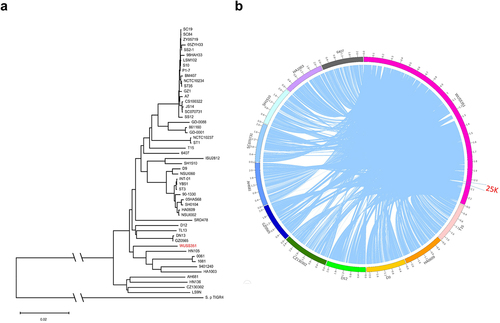
Figure 3. The T7SS of strain WUSS351. (a) Comparison of the sequence of the T7SS clusters between strains WUSS351 and GZ0565. (b) Results of competition experiments performed with WUSS351 or ΔT7SS as attacker strains and ΔT7SS as the prey strain. The number of the final prey cells was determined at 8, 18, and 32 h by serially diluting cultures onto LB+Spc agar plates. (c) Results of competition experiments performed with WUSS351 or ΔT7SS as attacker strains and S. suis strain P1/7-Rif as the prey strain. The number of the final prey cells was determined at 6, 8, and 10 h by serially diluting cultures onto LB+Rif agar plates. The data in the graphs represent the means ± SEM, and asterisks indicate pairs of significantly different values (n = 3, *** indicates p <0.001, * indicates p <0.05, two-tailed unpaired t test).
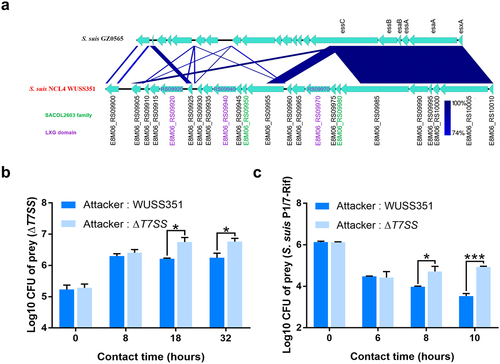
Figure 4. Lcn351 targets B. subtilis and S. suis. (a) The genetic organization of the lactococcin cluster (blue) and Lcn351 (yellow) in strain WUSS351. (b) The secreted proteins extracted from strain WUSS351 expressing Lcn351 fused with a SPA tag from its own promoter of the cluster (pSET2-lcn351) or control plasmid pSET2 were probed with FLAG or GroEL antibodies. (c) Alignment of the amino acid sequences of Lcn972 family bacteriocins: SP_0109 of S. pneumoniae strain TIGR4, SAP019 of S. aureus strain N315, Lcn972 of L. lactis strain IPLA 972, K710_0693 of S. iniae strain SF, and Lmo2776 of L. monocytogenes strain EGD-e. (d) The relative growth rate of different bacteria calculated after 6 h of incubation with supernatants of WT or Δlcn351. The relative growth rate of B. subtilis 1.460 (e) and S. suis P1/7 (f) after 6 h of incubation in the presence of Lcn351 peptide (10 or 20 mg/mL) or H2O; the relative growth rate was compared with Lcn351 group. The graph shows the means ± SEM; asterisks indicate pairs of significantly different values (n = 3, ** indicates p <0.01, * indicates p <0.05, ns indicates not significant, two-tailed unpaired t test).
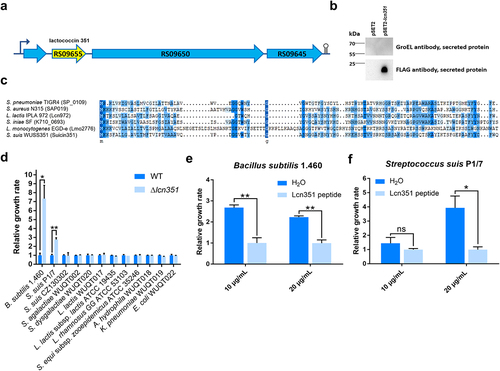
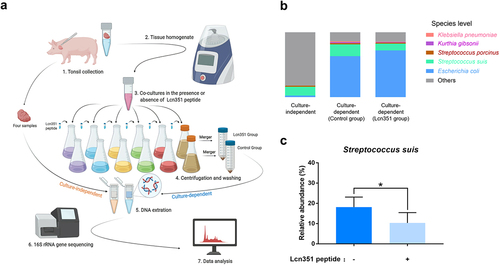
Figure 5. Lcn351 targets S. suis in pig tonsils microbiota. (a) A schematic diagram of the culture-independent and culture-dependent pig tonsillar microbiome analysis. Please refer to the Materials and methods section for the detailed protocols. (b) The relative abundance of species in the culture-independent and culture-dependent communities of pig tonsils. (c) The relative abundance of S. suis in the culture-dependent community of pig tonsils. The data in the graphs represent the means ± SEM, and asterisks indicate pairs of significantly different values (n = 4, * indicates p < 0.05, two-tailed unpaired t test).
Supplemental Material
Download Zip (1.8 MB)Data availability statement
The complete genome sequence of strain WUSS351 was deposited in GenBank and received the accession number NZ_CP039462.1. The data supporting this study’s findings are available from the corresponding author upon reasonable request.
