Figures & data
Table 1. Primers used in this study
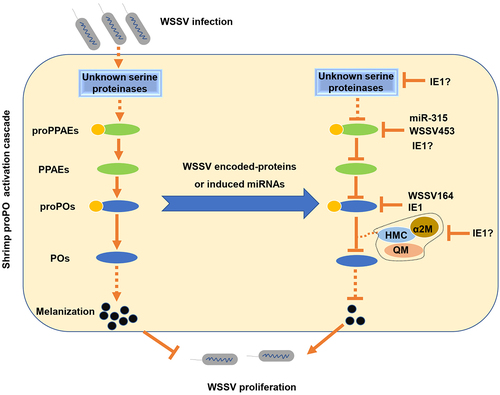
Figure 1. Identification of the host cellular proteins potentially interacting with WSSV IE1. (a) and (b) Expression of recombinant GST and GST-IE1 proteins. The GST or GST-IE1 expression plasmid transformed Escherichia coli cells were induced using 0 or 1 mM IPTG and then lysed for SDS-PAGE analysis. (c) SDS-PAGE analysis of the purified recombinant GST and GST-IE1 proteins. (d) GST pull-down and LC-MS/MS analysis of the candidate IE1-interacting proteins. The GST and GST-IE1-bound GSH magnetic beads were incubated with the hemocytes lysate supernatant (HLS). After incubation and wash, the bound proteins on the beads were eluted and then subjected to SDS-PAGE and silver staining analysis. The differential bands were excised from the gel and used for LC-MS/MS analysis. M: protein marker.
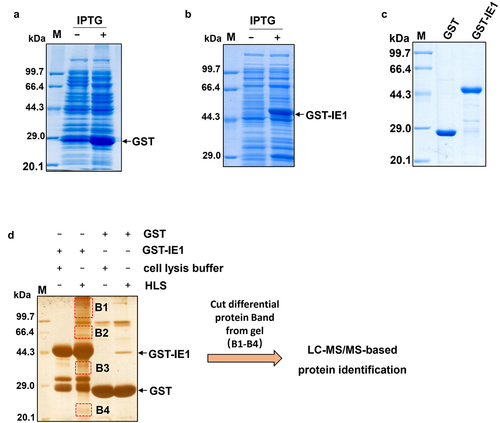
Figure 2. Functional classification analysis of the potential IE1-interacting proteins. (a) and (b) GO and KEGG pathway analysis of the identified IE1-interacting proteins. The GO and KEGG pathway entry ID of each identified IE1-interacting proteins were retrieved from our in-house P. vannamei transcriptome data, and then subjected to GO and KEGG pathway analysis using WEGO program and KEGG pathway database.
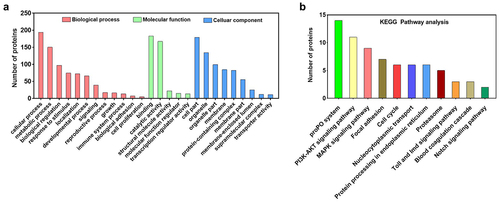
Figure 3. IE1 binds to the Ig-like domain of PvproPO and PvHMC. (a) Schematic comparison of PvproPO1, PvproPO2 and PvHMC in protein sequence identity and functional domains. (b-e) Co-IP analysis of the interactions between IE1 and PvproPO/PvHMC. The FLAG-tagged PvproPo1/pvpropo2/pvhmc/pvpropo1-Ig-like protein was co-expressed with the V5-tagged IE1 or EGFP (negative control) in High-five cells. At 48 h post-transfection, cells were harvested and lysed. The whole cell lysate (Input samples) were obtained by centrifugation and incubated with anti-FLAG M2 magnetic beads. The bound proteins (IP samples) were eluted from the beads using 3×FLAG peptide. Finally, the input (2%) and IP samples were subjected to western blot analysis with the indicated antibodies.
Table 2. Summary of the identified proPO system proteins that potentially binds IE1
Figure 4. IE1 inhibits PO activity and promotes viral replication in shrimp hemocytes. (a) Workflow of RNAi of IE1 during WSSV infection. (b-c) Knockdown efficiency analysis of IE1. hemocytes in dsRNA-IE1 or dsRNA-EGFP-injected shrimp were collected and used to prepare cDNA and protein lysates for qPCR and western blot analysis. (d) Detection of WSSV genes after IE1 silencing by relative qPCR. The mRNA expression of IE1 (WSV069), WSV051, WSV079, WSV514, ICP11 (WSV230), and VP28 (WSV421) was normalized to PvEF1α and calculated using the 2−ΔΔCT method. (e) Detection of viral loads by absolute qPCR. The genomic DNA was extracted from hemocytes in IE1 or EGFP-silenced shrimp, followed by absolute qPCR analysis, the WSSV copy number in 1 ng of shrimp genomic DNA was then calculated. (f) Determination of the PO activity after IE1 suppression. The hemocyte lysate supernatant (HLS) prepared from IE1 or EGFP-silenced shrimp were incubated with L-DOPA. After reaction for 30 min at room temperature, the absorbance at 490 nm was monitored, and the PO activity was determined as ΔA490/mg total protein/min. (g) Effect of recombinant IE1 protein on the PO activity. The HLS was incubated with recombinant GST-IE1 (0.2 μg or 2 μg) or the equal amount of GST protein (negative control) for 5 min at room temperature. After further reaction with L-DOPA, the PO activity was then determined by measuring the absorbance at 490 nm. All the experiments were carried out in independent triplicates, and the data were shown as Mean ± SD. The significance difference was analyzed using the student’s t-test (*p <0.05, ** p <0.01).
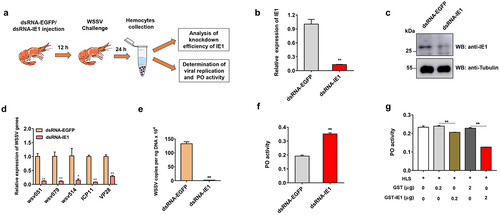
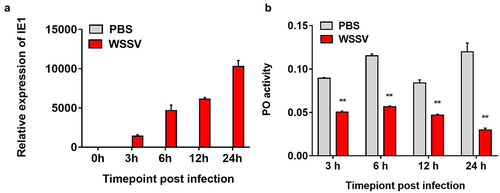
Figure 5. WSSV suppresses the shrimp PO activity at the early stage of infection. (a) qPCR analysis of IE1 expression during WSSV infection. (b) Detection of PO activity after WSSV infection. The shrimp was divided into two group, and then intramuscularly injected with WSSV or PBS (negative control). At 0, 3, 6, 12 and 24 h post-infection, the hemocytes per group were collected and used for qPCR and determination of PO activity. The data referred to the means ± SD of triplicate assays (**, p <0.01).
Supplemental Material
Download Zip (47.8 KB)Data availability statement
All the data supporting the findings of this study are available within the article and its supplementary materials.