Figures & data
Figure 1. WOR1 overexpression suppresses fitness defects of hog1 mutants.
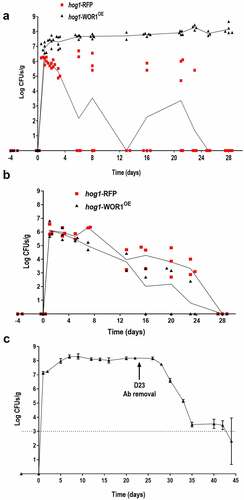
Figure 2. Competition of hog1-WOR1OE cells with wild-type cells.
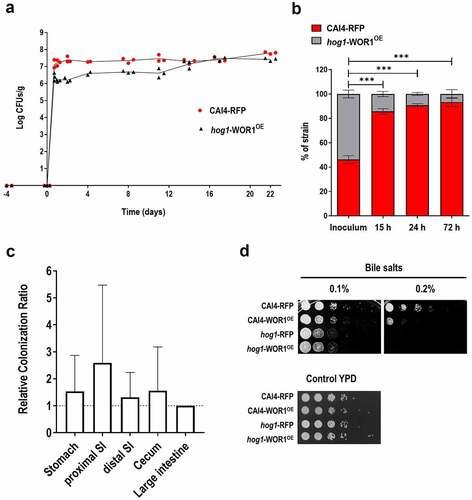
Figure 3. Effect of overexpression of WOR1 in hog1 cells adhesion.
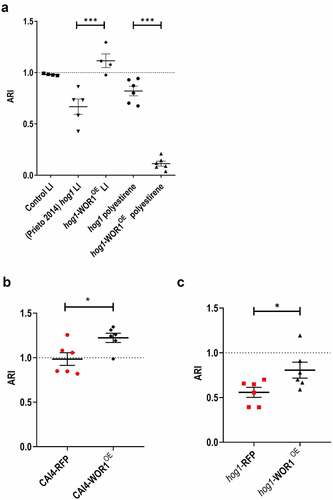
Figure 4. Role of Als3 in fitness.
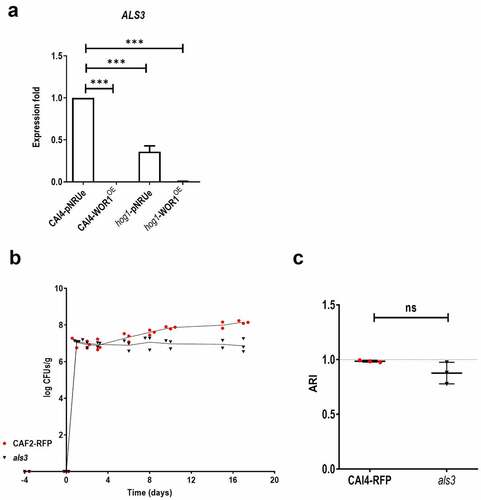
Figure 5. Effect of Wor1 overproduction in hog1 morphogenesis.
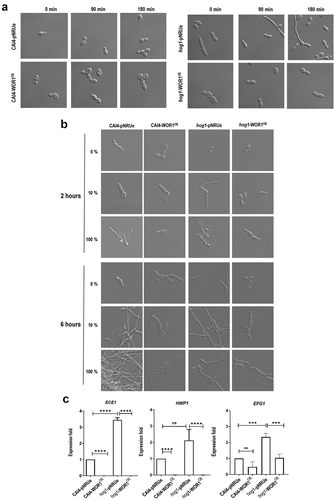
Figure 6. Determination of phospholipase and protease activity in WOR1OE cells.
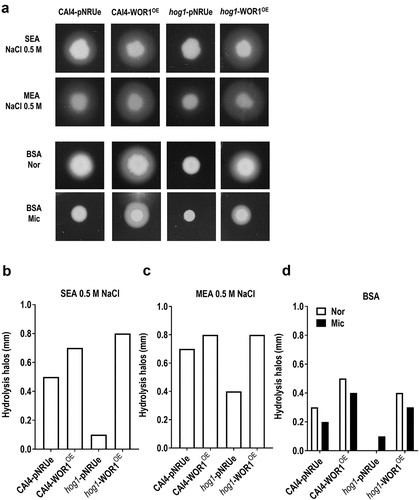
Figure 7. Effect of Wor1 overproduction in mouse viability (in a systemic candidiasis model).
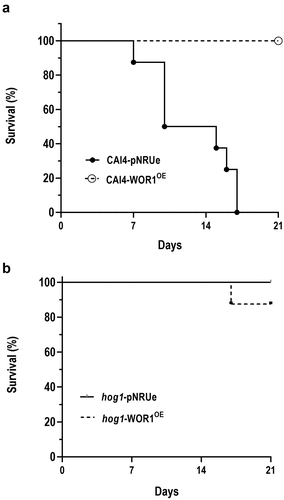
Table 1. Candida albicans strains used in this work.
Supplemental Material
Download Zip (1.1 MB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
