Figures & data
Figure 1. Screening for R. anatipestifer OMPs that interact with dVn. (a) SDS-PAGE detection of purified dVn. Lanes: M, molecular weight marker; 1, purified dVn. (b) OMPs of R. anatipestifer RA-YM were detected by SDS-PAGE and mainly were in the range of 20–120 kD. (c) Identification (by far-western blot) of OMPs of R. anatipestifer RA-YM that interact with dVn. (d) Pull-down with dVn of OMPs of the RA-YM strain by SDS-PAGE. Lanes: 1, purified dVn; 2, dVn pull down of OMPs; 3, negative control. (e) ELISA validation of His-tagged OMPs that interact with dVn. (f) Western blot validation of His6-OMP76 pull down using Strep-dVn. Three independent experiments were conducted and a representative experiment is shown here. The nature of data points belongs to biological repetition.
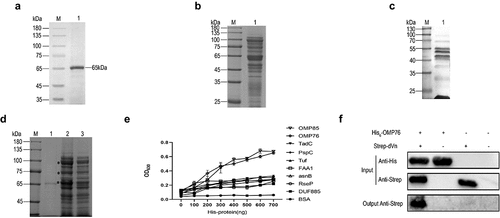
Figure 2. Expression of truncated OMP76 and screening for the OMP76 domain that interacts with dVn. (a) Detection of the immunogenicity and reactogenicity of OMP76 with R. anatipestifer RA-YM whole bacteria immune serum by Western blotting. The antibody used was a duck anti-R. anatipestifer RA-YM strain whole protein immune serum prepared in our laboratory, and the secondary antibody was HRP*PCAB rabbit anti-duck IgY (IgG)(H+L) (Cellwaylab, Luoyang, China). (b) the tertiary structure of OMP76 predicted by AlphaFold. Red, helix; yellow, β-strand; green, loop. (c) the position of truncated fragments derived from full-length OMP76. (d) ELISA analysis of the interaction of truncated segments of OMP76 with dVn. (e) Pull-down assay of the interaction of truncated segments L5 and L11, and knockdown fragment OMP76Δ307–315 with dVn by Western blotting. ELISA and pull-down assays were repeated three times and representative experiments are shown here. The nature of data points belongs to biological repetition.
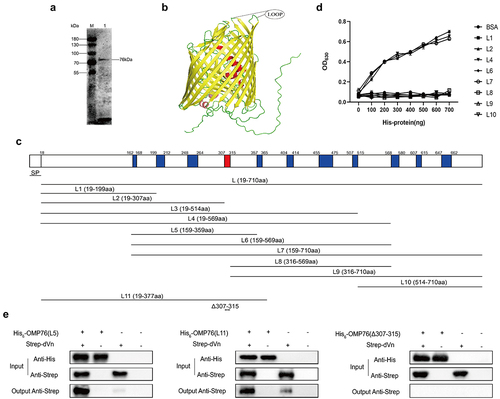
Figure 3. Validation of the surface localisation of OMP76 and the ability of OMP76 to mediate bacterial binding of dVn. (a) FACS analysis of R. anatipestifer RA-YM and R. anatipestifer RA-YMΔOMP76 binding to OMP76 pAb. (b) FACS analysis of E. coli BL21(DE3)/BL21(DE3)/Pet−16b−16b-OMP76 and E. coli BL21(DE3)/pET−16b binding to OMP76 pAb. (c) Western blot assay of binding of the RA-YM and RA-YMΔOMP76 strains to dVn in normal duck serum and to purified dVn protein. (d) Western blot assay of E. coli BL21(DE3)/BL21(DE3)/Pet−16b−16b-OMP76 and E. coli BL21(DE3)/pET−16b binding to dVn in normal duck serum and to purified dVn protein. (e) FACS analysis of binding of the RA-YM and RA-YMΔOMP76 strains to purified dVn protein. (f) FACS analysis of binding of E. coli BL21(DE3)/BL21(DE3)/Pet−16b−16b-OMP76 and E. coli BL21(DE3)/pET−16b to purified dVn protein. A representative image of three independent experiments is shown. The nature of data points belongs to biological repetition.
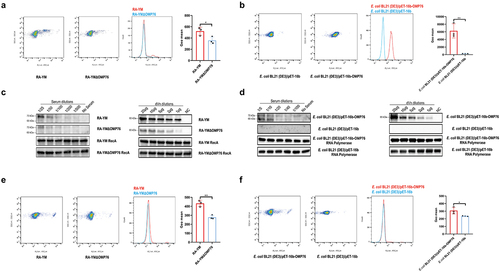
Figure 4. Western blot detection of C9 multimers deposited on the bacterial cell surface. (a) Western blotting assay of C9 deposition on R. anatipestifer RA-YM and R. anatipestifer RA-YMΔOMP76 when incubated in 5% NDS for the indicated time periods. (b) Western blotting assay of C9 deposition on E. coli BL21(DE3)/BL21(DE3)/Pet−16b−16b-OMP76 and E. coli BL21(DE3)/pET−16b when incubated in 5% NDS for the indicated time periods. (a) and (b) were repeated three times and representative images of these independent experiments are shown.
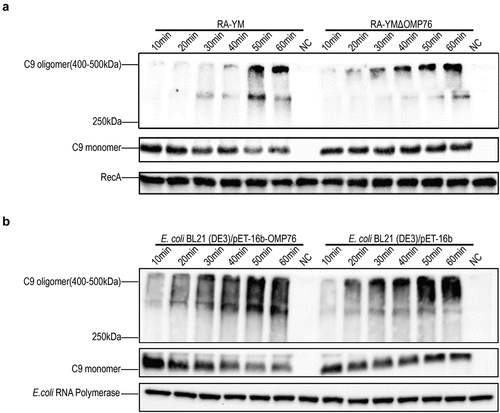
Figure 5. Serum survival assays. (a) Serum survival of R. anatipestifer RA-YM, R. anatipestifer RA-YMΔOMP76, and R. anatipestifer RA-YMCΔOMP76 in 10% NDS. (b) Serum survival of E. coli BL21(DE3)/BL21(DE3)/Pet−16b−16b-OMP76 and E. coli BL21(DE3)/pET−16b in 2% NDS. Panels (a) and (b) also show the survival of the RA-YM, RA-YMΔOMP76, and RA-YMCΔOMP76 strains and E. coli BL21(DE3)/BL21(DE3)/Pet−16b−16b-OMP76 and E. coli BL21(DE3)/pET−16b in HIS at the same serum dilution concentrations thereby assessing complement activity in serum. The data were analysed by two-way ANOVA and error bars represent standard deviations.
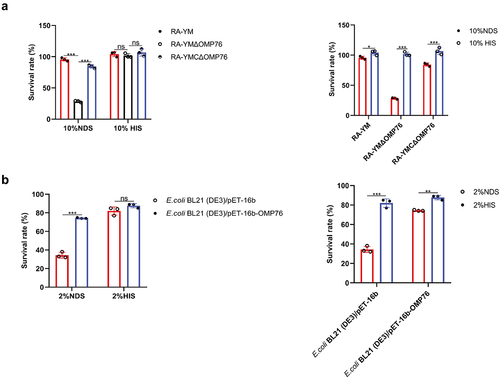
Figure 6. Adhesion and invasion assays of R. anatipestifer RA-YM, R. anatipestifer RA-YMΔOMP76, and R. anatipestifer RA-YMCΔOMP76. (a) Adhesion assays of the RA-YM, RA-YMΔOMP76, and RA-YMCΔOMP76 strains to Vero cells. (b) Invasion assay of the RA-YM, RA-YMΔOMP76, and RA-YMCΔOMP76 strains to Vero cells. All data are mean values of three independent experiments that were analysed by one-way ANOVA. Error bars represent the standard deviations.
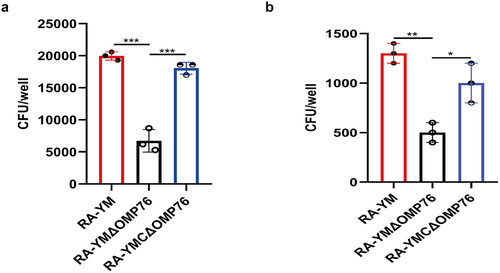
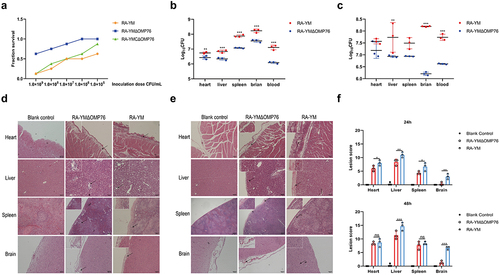
Figure 7. Determination of blood and tissue load of ducklings infected with R. anatipestifer RA-YM or R. anatipestifer RA-YMΔOMP76. (a) Mortality of ducklings infected with R. anatipestifer RA-YM, R. anatipestifer RA-YMΔOMP76, and R. anatipestifer RA-YMCΔOMP76. (b) Bacterial load in the heart, liver, spleen, and brain of Cherry Valley ducks infected with the RA-YM or RA-YMΔOMP76 strains for 24 h. (c) Bacterial load in the heart, liver, spleen, and brain of Cherry Valley ducks infected with the RA-YM or RA-YMΔOMP76 strains for 48 h. Two-way ANOVA was used to assess differences in bacterial numbers in different tissues. *P≤0.05; **P≤0.01; ***P≤0.001. (d) Histopathological analysis of ducklings infected with the RA-YM or RA-YMΔOMP76 strains for 24 h. (e) Histopathological analysis of ducklings infected with the RA-YM or RA-YMΔOMP76 strains for 48 h. (f) Lesion score of the heart, liver, spleen, and brain of Cherry Valley ducks infected with the RA-YM or RA-YMΔOMP76 strains for 24h and 48 h. Two-way ANOVA was used to assess differences in lesion score in different tissues. *P≤0.05; **P≤0.01; ***P≤0.001. The nature of data points belongs to biological repetition.
Supplemental Material
Download MS Word (5 MB)Data availability statement
The original contributions presented in the study are included in the article and Supplementary Materials. Further inquiries can be directed to the corresponding author. The mass spectrometry raw files have been deposited to ProteomeXchange (https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD039753) with the accession number PXD039753.
