Figures & data
Figure 1. Illustrative Rab-related pathways used by viruses. I. Rab proteins involved in entry pathways are pictured on the right side of the diagram. Numerous viruses utilize the endocytic pathway to enter cells, and may deliver their capsids to the cytoplasm through Rab5+ or Rab7+ endosomes. II. Rab proteins play critical roles in the assembly and egress of enveloped viruses from cells. Rab11 and Rab 11 FIPs (adaptor proteins) have been implicated in RSV, CMV, and HIV assembly. Rab9 plays a role in late events for several viruses by an as yet unclear mechanism. III. Viral glycoprotein trafficking follows Rab-directed vesicular trafficking pathways, and may involve both outward movement to the plasma membrane and/or endocytosis to intracellular sites of assembly.
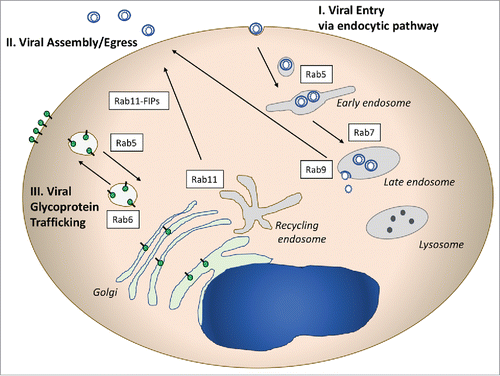
Figure 2. Herpesviruses and Rab Proteins. (A) Herpes simplex virus Type 1 (HSV-1) assembly and Rab proteins. HSV-1 utilizes Rab6-dependent TGN-to-PM transport of its glycoproteins, which are subsequently endocytosed in a Rab5-dependent process. Rab11 is essential for re-envelopment of capsids. (B) Human Cytomegalovirus (HCMV) utilization of Rab protein trafficking. Pictured schematically is the CMV assembly compartment, where a series of Rab proteins are recruited and contribute to assembly. Rab6 interacts with the adaptor BicD1 to bring the tegument protein pp150 to the assembly compartment in a dynein-dependent transport process.
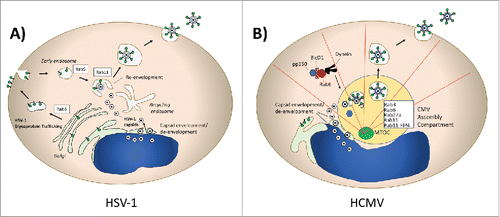
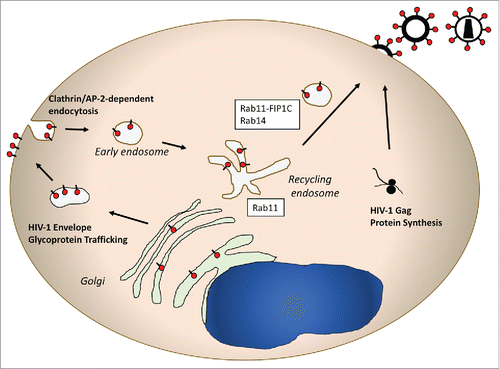
Figure 3. Schematic representation of HIV-1 envelope trafficking and assembly. This pathway features a prominent role for Rab14 and Rab11-FIP1C in directing the viral envelope to the plasma membrane site where the Gag lattice forms. Note that in a manner similar to HSV-1 glycoprotein trafficking, the HIV envelope glycoprotein is transported from TGN-to-PM, followed by endocytosis to the endosomal recycling compartment. From there, Rab11-FIP1C and Rab14 are required for outward sorting to the particle assembly site on the PM.